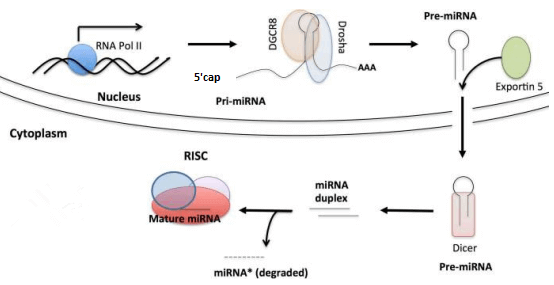
3. Nomenclature
A.The system name of a miRNA generally consists of three parts: species, category, serial number, and they are connected by short lines.
B. Human, mouse, and rat are the three most popular organisms for researchers, and they are named with the prefixes hsa, mmu, and
rno, respectively.
C.Categories can distinguish the form of miRNA. mir refers to a pre-miRNA or pri-miRNA, miR represents a mature miRNA.
D. The serial number represents the order of miRNA discovery. In general, the earlier the discovery, the smaller the number.
E. Sometimes miRNA names are suffixed, such as hsa-miRNA-19a and hsa-miRNA-19b, indicating that the two miRNA sequences are
highly homologous; hsa-miRNA-19-1 and hsa-miRNA-19-2 indicate that their precursor sequences are different, but the mature miRNA sequences are identical; hsa-miRNA-19-5p and hsa-miRNA-19-3p
indicate that the two miRNAs are processed from the 5' and 3' arms of the same precursor. (Sometimes also expressed by hsa-miRNA-19-s and hsa-miRNA-19-as).
F.The miRNAs found before the naming convention, such as lin-4, let-7, still retain the original name.
4.1 Biological roles
miRNAs are involved in various processes such as plant and animal growth and development, cell differentiation, cell proliferation and
apoptosis, hormone secretion, and tumor formation.
When the first miRNA was discovered, scientists realized that this small molecule has the effect of inhibiting and regulating genes.
What mechanism does miRNA repress other genes, leading to inhibition of protein synthesis, and further regulation of cell differentiation and tissue development? After more than two decades of in-depth
research, scientists have found the possible mechanism by which miRNAs regulate genes.
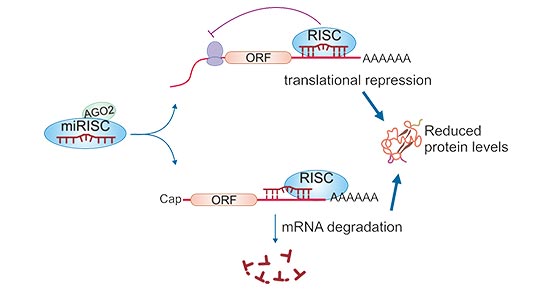
The mature miRNA in the RISC complex binds to the 3'UTR of the target mRNA (messenger RNA) through a 2 to 8 nucleotide sequence at
the 5' end, thereby suppressing translation or degradation of mRNA. This process is also called post-transcriptional gene silencing (PTGS). The 2 to 8 nucleotide sequence at the 5' end of the miRNA is
called "seed" region; the sequence in the 3'-UTR of the target mRNA acts on the miRNA is called a miRNA reaction element (MRE). But some studies have shown that it also occurs in 5'-UTR, even in protein
coding sequences. For mammals, base pairing is always imperfect, such as pairing between let-7 and lin-41.
When the seed region of the miRNA is completely or nearly completely complementary to the MRE of the target mRNA, the mRNA is
degraded; while their binding is incomplete, protein synthesis is inhibited. Both of these combinations can result in a decrease in the overall level of the protein, thus leading to gene regulation.
One miRNA can bind hundreds of target mRNAs in sequence, and a single mRNA can be targeted by several miRNAs.
Figure 2: Mechanism of action of miRNA
(sourcing: http://dmd.aspetjournals.org/content/44/3/308)
4.2 Medical roles
miRNA can also serve as a diagnostic or therapeutic target for one or more diseases. When miRNAs target tumor suppressor mRNA,
their roles are oncomiRs; on the one hand, they act as tumor suppressors by targeting mRNAs encoding oncoproteins.
Deregulation of miRNAs in disease states can be exploited as potential therapeutic agents by miRNA replacement therapy using miRNA
mimics or by inhibition of miRNA function by anti-miRNA.
Many preclinical studies using various disease models have tested the use of these next-generation therapies, and some miRNA-based
therapies have entered clinical testing.
5. The diseases caused by micro ribonucleic acid disorders
miRNA dysregulation has proved to be related to many diseases, especially cancers. A notable feature of cancer is the disorder of gene
expression. This suggests that miRNAs affect tumor growth, invasion, metastatic ability, and angiogenesis, etc. miRNAs can act as oncogenes or tumor suppressors.
Here list some cancers associated with miRNAs dysregulation.
|
miRNA
|
Cancer Type
|
Suppressed Gene
|
|
Tumorigenesis and Apoptosis
|
|
miR-423
|
Breast cancer
|
Human progesterone receptor(PR)
|
|
miR-211
|
Mammary carcinoma, B cell lymphoma
|
CHOP
|
|
miR-370, miR-1180, miR-1236
|
Bladder cancer
|
p21
|
|
miR-205
|
Prostate cancer
|
IL24 IL32
|
|
miR-124
|
Breast and ovarian cancer
|
p27
|
|
miR-2478
|
Breast cancer
|
TGFβ1
|
|
miR-138
|
Prostate cancer
|
β-catenin
|
|
miR-877
|
Bladder cancer
|
p16
|
|
miR-6734
|
Colon Cancer
|
p21
|
|
Metastasis and Angiogenesis
|
|
miR-10a
|
Breast cancer
|
Hoxd4
|
|
miR-9
|
Non-small-cell lung carcinoma
|
MALAT-1
|
|
miR-205
|
Prostate cancer IL24
|
IL32
|
|
miR-337
|
Neuroblastoma
|
Matrix metalloproteinase
|
|
miR-584
|
Human neuroblastom
|
Matrix metalloproteinase
|
|
miR-558
|
Neuroblastoma
|
Heparanase
|
|
miR-337
|
Gastric cancer
|
Matrix metalloproteinase
|
|
miR-215
|
Malignant gliomas
|
PCDH9
|
|
miR-2478
|
Breast cancer
|
TGFβ1
|
(sourcing: https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-018-0765-5)
In conclusions, miRNA is a non-coding RNA with 21 to 23 nucleotides in length. Although it is small, it plays an essential regulatory role
in life activities.
The number of miRNA genes being discovered is increasing, and new mechanisms of action may reveal possible new therapeutic
strategies. The discovery and development of miRNA inhibitors or miRNA mimics as new drugs will provide new hopes for combating cancer. All in all, the deepening of small molecule miRNA research will
help us uncover more of the mysteries of life.




Comments
Leave a Comment