Proteins play important roles in biological processes, drug discovery and development, disease diagnosis and treatment, and bioengineering and biotechnology. The importance and wide application of proteins make almost all scientific research involves proteins. Protein-based studies usually involve complicated procedures including protein expression, detection, purification, localization, interaction, etc, which are often laborious and non-generic.
Adding specific protein tags to the target protein makes it easily combined with affinity resin or detection antibodies, achieving efficient purification and detection. Protein tags can also be used to track protein localization and interactions, as well as regulate protein expression and stability.
This article provides a comprehensive overview of protein tags, listing commonly used tags, categorizing them into affinity, epitope, and fluorescent tags, elucidating each tag's characteristics, and discussing how to tag a protein and how to select an appropriate tag for a certain protein.
1. What Is A Protein Tag?
A protein tag is a peptide or protein that is genetically fused to either the N- or C-terminal of the protein of interest during the cloning step for a specific purpose. These tags vary in size, ranging from just a few amino acid residues to full-length proteins or domains.
2. Functions of Protein Tags
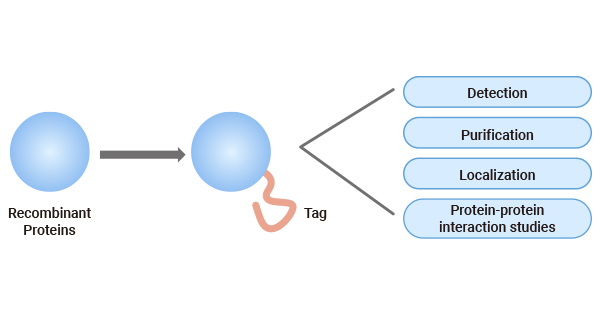
As a type of important tool in biological research, protein tags confer the target protein with additional properties such as affinity, solubility, and fluorescence, and thus widely used in multiple fields, including protein purification, protein detection and quantification, solubility enhancement, protein yield improvement, subcellular protein localization, protein tracing, and protein-protein interaction research [1].
Figure 1. Main Functions of protein tags
Protein tags are useful in recombinant protein production. Different protein tags may execute distinct functions but may be overlapping:
Enhance protein affinity and solubility [2,8];
Increase the expression of target proteins [9, 13];
Facilitate the isolation and purification of target proteins;
Protect target proteins from degradation [10-12];
Augment the folding ability of target protein [14].
Protein tags can be recognized by specific antibodies, affinity resins, or other binding partners, enabling researchers to manipulate and study the tagged protein with greater ease and precision.
3. Common Protein Tags
There are a variety of available protein tags in the market. According to their properties, protein tags can be roughly divided into affinity tags, epitope tags, and fluorescent tags. The following table lists commonly used protein tags, protein tag comparisons, as well as protein tag advantages and disadvantages.
Table 1. Commonly used protein tags
| Tag Classification |
Feature |
| Affinity tags |
His tag |
Sequence or Mass: 6*His: HHHHHH (o.84 kDa)
Fused Position: C- or N-terminal
Application: Affinity and purification
Strengths: Small size has little effect on the function of the target protein and do not form dimer and thus can be used for downstream analyse;
Can purify under native and denaturing conditions;
Low immunogenicity;
Can be eluted under mild conditions by imidazole competition or low pH
Weaknesses: Non-specific binding to nickel resin |
| GST (Glutathione-S-transferase) |
Sequence or Mass: 211 amino acid residues, 26 kDa
Fused Position: C- or N-terminal
Application: Purification and solubility enhancement
Strengths: High affinity for glutathione resin;
Protect from proteolysis of the target protein and improve its stability;
Can be eluted under mild and non-denaturing conditions, thus preserving the antigenicity and biological activity of the protein
Weaknesses: Unsuitable for tagging multimeric protein complexes due to its four solvent exposed cysteines that can provide a significant oxidative aggregation |
| MBP (Maltose-binding protein) |
Sequence or Mass: 396 amino acid residues, 42.5 KDa
Fused Position: C- or N-terminal
Application: Detection, purification, increased expression and solubility
Strengths: Can reduce the degradation of the target protein and enhance the solubility and folding of eukaryotic proteins in prokaryotes;
Can be easily detected through immunoassays
Weaknesses: Larger size may affect protein structure and function |
| TAP (Tandem affinity purification) |
Sequence or Mass: Variable
Fused Position: C- or N-terminal
Application: Protein purification
Strengths: Two-step purification can obtain highly pure protein with minimal background;
Mild elution conditions do not affect fusion properties; Protein of interest remains in native form;
Use of protease unnecessary for elution;
Over 30 TAP variants available
Weaknesses: A multi-step purification process may be time-consuming
Use of multiple affinity resins and reagents may make it more expensive |
| Strep-II (Streptavidin binding peptide) |
Sequence or Mass: WSHPQFEK (1.06 kDa)
Fused Position: C- or N-terminal, or within the target protein
Application: Detection, purification and immobilization
Strengths: Short, linear recognition motif;
Matrix regenerable;
Useful for purification under anaerobic conditions, eukaryotic cell surface display, and immobilization to streptavadin-coated surfaces;
Elution from streptavidin columns with biotin derivates under gentle conditions
Weaknesses: Specific binding conditions may be unsuitable for some fusions |
| Calmodulin binding peptide (CBP) |
Sequence or Mass: 26 amino acid residues, 4 kDa
Fused Position: C- or N-terminal
Application: Protein purification, protein-protein interaction study
Strengths: CBP tightly binding to calmodulin allows efficient purification under mild conditions;
Addition of calcium-chelating allows the single step elution of the target protein under mild conditions;
Weaknesses: Binding between CBP and calmodulin requires calcium ions, limiting purification under calcium-free conditions;
CBP-tagged protein purification may involve the use of specialized calmodulin resin or reagents, increasing overall cost |
| CBD (Intein-chitin binding domain) |
Sequence or Mass: 51 amino acid residues
Fused Position: C- or N-terminal
Application: Purification
Strengths: The intein domain enables precise cleavage at the intein-CBD junction, allowing tag removal without additional proteases;
Facilitate high-affinity binding to chitin or chitin derivatives, enabling efficient protein purification
Weaknesses: Intein-mediated cleavage may produce impurities in purified samples |
| Halo Tag (mutated dehalogenase) |
Sequence or Mass: ~300 amino acid residues, 33 KDa
Fused Position: C- or N-terminal
Application: Purification, and increase solubility and expression
Strengths: Can covalently and specifically bind to multiple synthetic reporter groups and affinity ligands, enabling Halo-tagged proteins to be used for the detection of affinity binding or solid-phase immobilized target proteins
Weaknesses: A different ligand must be purchased for each different experiment;
Large size and stringent wash conditions may affect properties of the target proteins |
| SUMO |
Sequence or Mass: ~100 amino acid residues, 12 kDa
Fused Position: N-terminal
Application: Increase solubility and expression
Strengths: Allow for cellular protein-protein interaction studies; Available for bacterial, yeast, insect, and mammalian expression systems;
Natural SUMO protease can highly specifically cleavage the tertiary SUMO structure;
Desumoylases and SUMO tags contain 6*His tag to make their removal efficient
Weaknesses: Large size may affect the folding and function of the target proteins;
May sometimes conjugate to unintended target proteins, leading to off-target effects |
| Protein A (Staphylococcal protein A) |
Sequence or Mass: 280 amino acid residues
Fused Position: N-terminal
Application: Purification and solubility enhancement
Strengths: Proteolytically stable;
May increase solubility of the target proteins
Weaknesses: Purification does not give high yields;
Large tag size and/or low pH elution may irreversibly affect protein properties;
Matrix is of limited reusability |
| Epitope tags |
FLAG |
Sequence or Mass: DYKDDDDK (1 KDa)
Fused Position: N-terminal
Application: Affinity, purification, and protein-protein interaction study [3]
Strengths: High specificity for FLAG antibody-based purification, detection, and identification;
Posses an intrinsic enterokinase cleavage site (DDDDK) at its C-terminus, allowing its complete detachment from the target protein [4];
Expression efficiency is higher in eukaryotic expression systems
Weaknesses: Antibody purification does not give high yields;
Low pH elution may irreversibly affect protein properties;
Matrix is of limited reusability |
| HA (Human influenza hemagglutinin) |
Sequence or Mass: 31 amino acid residues
Fused Position: C- or N-terminal
Application: Protein purification and detection
Strengths: Anti-HA antibodies specific;
Useful in mammalian expression systems;
Suitable for multiple downstream analyse, including WB, IP, IF, ELISA, and FC
Weaknesses: Antibody purification does not give high yields;
Low pH elution may irreversibly affect protein properties;
Matrix is of limited reusability |
| V5 (Bacteriophage V5 epitope) |
Sequence or Mass: GKPIPNPLLGLDST (1.4 kDa)
Fused Position: C-terminal
Application: Protein detection and localization, flow cytometry, affinity chromatography purification, protein research, and quantitative analysis
Strengths: Short, linear recognition motif;
Antibody specific in bacterial lysates;
Often used in combination with His-tag for protein purification
Weaknesses: Exist some cross-reactivity in mammalian lysates |
| c-Myc |
Sequence or Mass: EQKLISEEDL (1.2 kDa)
Fused Position: C- or N-terminal
Application: Detection and purification
Strengths: Short, linear recognition motif;
Frequently used for western blots, IP, co-IP, IF, flow -cytometry
Weaknesses: Myc-tagged protein can not be used in human cells or tissue-related experiments due to the Myc tag being part of the human Myc gene;
Low pH elution may irreversibly affect protein properties;
Matrix is of limited reusability |
| Fluorescent tags |
GFP (green fluorescent protein) |
Sequence or Mass: 238 amino acid residues, 25 kDa
Fused Position: C- or N-terminal
Application: Protein cellular localization, cell physiological process monitoring, and detection of transgene expression in vivo
Strengths: Emit green light when illuminated with blue or UV light, achieving simple and intuitive observation;
Can detect directly with a fluorescence microscope, facilitating the real-time monitoring of the subcellular localization and expression of target proteins in living single cells in situ
Weaknesses: Some GFP fusions non-specifically targeted to nucleus; Very large tag or GFP dimerization may affect properties of the target proteins |
| Luciferase |
Sequence or Mass: 551 amino acide residues
Fused Position: N-terminal
Application: Protein detection
Strengths: Luminescent;
Can serve as a reporter immediately upon translation;
Useful for studies involving in situ hybridization, RNA processing, RNA transfection or coupled in vitro transcription/translation, protein folding, and imaging
Weaknesses: No more than five codons can be removed from the N- or C-term to maintain enzymatic activity;
Very large tag may affect properties of the target proteins |
| Others |
Avi tag |
Sequence or Mass: GLNDIFEAQKIEWHE (1.6 kDa)
Fused Position: C- or N-terminal
Application: Isolation, purification, and protein-protein interaction study
Strengths: Almost all proteins can be easily and efficiently biotinized at a unique Avi Tag site, both in vitro and in vivo;
Biotinylation conditions are quite mild and Avi-Tag is extremely highly specific;
Small size minimally affects the folding and function of the target protein
Weaknesses: Efficiency may vary depending on the accessibility of the Avi tag and the conditions of the biotinylation reaction |
4. How to Select An Appropriate Protein Tag
Selecting an appropriate protein tag depends on the specific requirements of the conducted experiment or application. Here are some factors to consider when choosing which protein tag to use in your research system:
Purpose of the fusion: Determine whether the purpose of adding protein tags is, for example, detection, purification, localization, or other. Generally, His-tag and Flag-tag are preferred for protein purification and Western blot, respectively. Flag, HA, and cMyc tags can be selected for immunoprecipitation, and GFP and CFP protein tags are used for immunofluorescence purposes.
Protein tag size: Whether the target protein needs a larger or smaller tag depending on its application. In theory, the larger the added protein tag, the greater the impact on the function and structure of the target protein. Small tags are useful for protein detection and antibody generation since they are less immunogenic than larger tags [5]. Common tags with low molecular weight include 6*His, Flag, HA, and c-Myc, which have only a few amino acids and rarely affect the structure and function of the protein.
Required production levels: Higher levels of protein production are needed in structural studies, which can be quickly attained by using a larger fusion tag with robust translational initiation signals, while the investigation of physiological interactions calls for lower production levels and smaller tags [1].
Tag location: Protein tags placed at the N-terminus or C-terminus of the target protein can promote different effects. Typically, N-terminal tagging is more beneficial than C-terminal tagging. N-terminal tags are friendly for efficient translation initiation, enabling fusion proteins to exploit the tag's efficient translation initiation sites. Most endoprotease cleavage at or near the C-terminus of their recognition sites leaves none or a few additional residues at the native N-terminal sequence of the target protein after N-terminal tags are removed [1,6].
However, If you do not have the exact protein structure or protein functional domain map, it is recommended to construct both N-terminal tagged and C-terminal tagged expression clones to test which one is more effective.
Protein tag removal: Every protein tag, whether large or small, may interfere with downstream functions of the target proteins. So, protein tag cleavage from the target proteins is necessary whenever it is feasible. To accomplish this, specific protease cleavage sites are placed in between the tag and the target protein. Site-specific proteases function in the cleavage site, releasing the target protein in its natural form. Two kinds of proteases endoproteases and exoproteases can be used to detach tags.
Table 2. Common endoproteases for tag removal [7]
| Proteases |
Source |
Cleavage site |
| TEV |
Tobacco etch virus protease |
ENLYFQ/G |
| Entk |
Enterokinase |
DDDDK/ |
| Xa |
Factor Xa |
IEGR/ |
| Thr |
Thrombin |
LVPR/GS |
| PreScission |
Genetically engineered derivative of human rhinovirus 3C protease |
LEVLFQ/GP |
| SUMO protease |
Catalytic core of Ulp1 |
Recognize SUMO tertiary structure and cleave at the C-terminal end of the conserved Gly-gly sequence in SUMO |
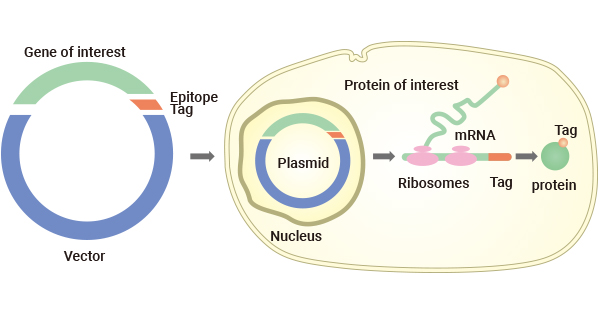
5. How to Tag A Protein
Once an appropriate protein tag has been selected, primers with sequences complementary to the upstream region of the gene of interest and the desired protein tag sequence are designed.
Amplify the target gene using PCR with the designed primers, incorporating the protein tag sequence at the 5'- or 3' -end based on the experiment goal.
Clone the PCR product into a suitable expression vector using restriction enzyme digestion and ligation or Gibson assembly.
After cloning, the resulting vector contains the target gene fused with the protein tag gene at either the N- or C-terminal. The tagged vector is transfected into a suitable host organism for protein expression. The presence of the specific tag enables efficient and thorough purification of the tagged protein, giving rise to highly pure proteins.
Due to their potential to impact the structure or activity of the target protein, protein tags are often removed from their fusion partners using highly sequence-specific proteases for downstream biological and functional studies.
Figure 2. Protein Tagging
6. Protein Tag-associated Products
CUSABIO provides many tag antibodies that are helpful for the recognition and detection of tagged proteins.
| Products |
Applications |
| 6*His Monoclonal Antibody |
ELISA, WB |
| Myc tag Monoclonal Antibody |
ELISA, WB, IF, IP |
| Flag Tag Monoclonal Antibody |
ELISA, WB, IF, IP |
| GFP Monoclonal Antibody |
ELISA, WB, IF, FC, IP |
| Sumo tag Monoclonal Antibody |
ELISA,WB |
| GST Monoclonal Antibody |
ELISA, WB, IF, FC, IP |
| MBP Monoclonal Antibody |
ELISA,WB |
| Avi-Tag Monoclonal Antibody |
ELISA,WB |
| CBP Tag Monoclonal Antibody |
ELISA,WB |
| V5-Tag Monoclonal Antibody |
ELISA, WB, IF, IP, FC |
| HA-Tag Monoclonal Antibody |
ELISA, WB, IF, IP, FC |
Conclusion
Protein tags genetically fused to a target protein have revolutionized detection and purification procedures, enabling researchers to study protein function, localization, and interactions with high precision. By understanding the diverse range of protein tags available, researchers can tailor their experiments to achieve optimal results.
References
[1] Malhotra, A. (2009). “Tagging for protein expression,” in Guide to Protein Purification, 2nd Edn, eds R. R. Burgess and M. P. Deutscher (San Diego, CA: Elsevier), 463, 239–258.
[2] Esposito, D., and Chatterjee, D. K. (2006). Enhancement of soluble protein expression through the use of fusion tags [J]. Curr. Opin. Biotechnol. 17, 353–358.
[3] Hopp T. P., Prickett K. S., et al. (1988). A short polypeptide marker sequence useful for recombinant protein identification and purification [J]. Biotechnology 6 1204–1210.
[4] Einhauer A., Jungbauer A. (2001). The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins [J]. J. Biochem. Biophys. Methods 49 455–465.
[5] Terpe K. (2003). Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems [J]. Appl. Microbiol. Biotechnol. 60 523–533.
[6] Waugh, D. S. (2005). Making the most of affinity tags [J]. Trends Biotechnol. 23, 316-320.
[7] Costa S, Almeida A, Castro A, Domingues L. Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system [J]. Front Microbiol. 2014 Feb 19;5:63.
[8] Maina C.V., Riggs P.D., et al. An Escherichia coli vector to express and purify foreign proteins by fusion to and separation from maltose-binding protein [J]. Gene. 1988;74:365–373.
[9] Hansted J. G., Pietikainen L., et al. (2011). Expressivity tag: a novel tool for increased expression in Escherichia coli [J]. J. Biotechnol. 155 275–283.
[10] Butt T. R., Edavettal S. C., et al. (2005). SUMO fusion technology for difficult-to-express proteins [J]. Protein Expr. Purif. 43 1–9.
[11] Nikaido H. (1994). Maltose transport-system of Escherichia coli: an ABC-type transporter [J]. FEBS Lett. 346 55–58.
[12] Kishi A., Nakamura T., et al. (2003). Sumoylation of Pdx1 is associated with its nuclear localization and insulin gene activation [J]. Am. J. Physiol. Endocrinol. Metab. 284 E830–E840.
[13] Ohana R. F., Encell L. P., et al. (2009). HaloTag7: a genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification [J]. Protein Expr. Purif. 68 110–120.
[14] Sacchetti, A., & Alberti, S. (1999). Protein tags enhance GFP folding in eukaryotic cells [J]. Nature Biotechnology, 17(11), 1046.
[15] Spriestersbach A, Kubicek J, et al. Purification of His-Tagged Proteins [J]. Methods Enzymol. 2015;559:1-15.
CUSABIO team. Protein Tags: A Comprehensive Overview. https://www.cusabio.com/c-20622.html




Comments
Leave a Comment