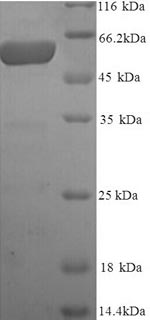
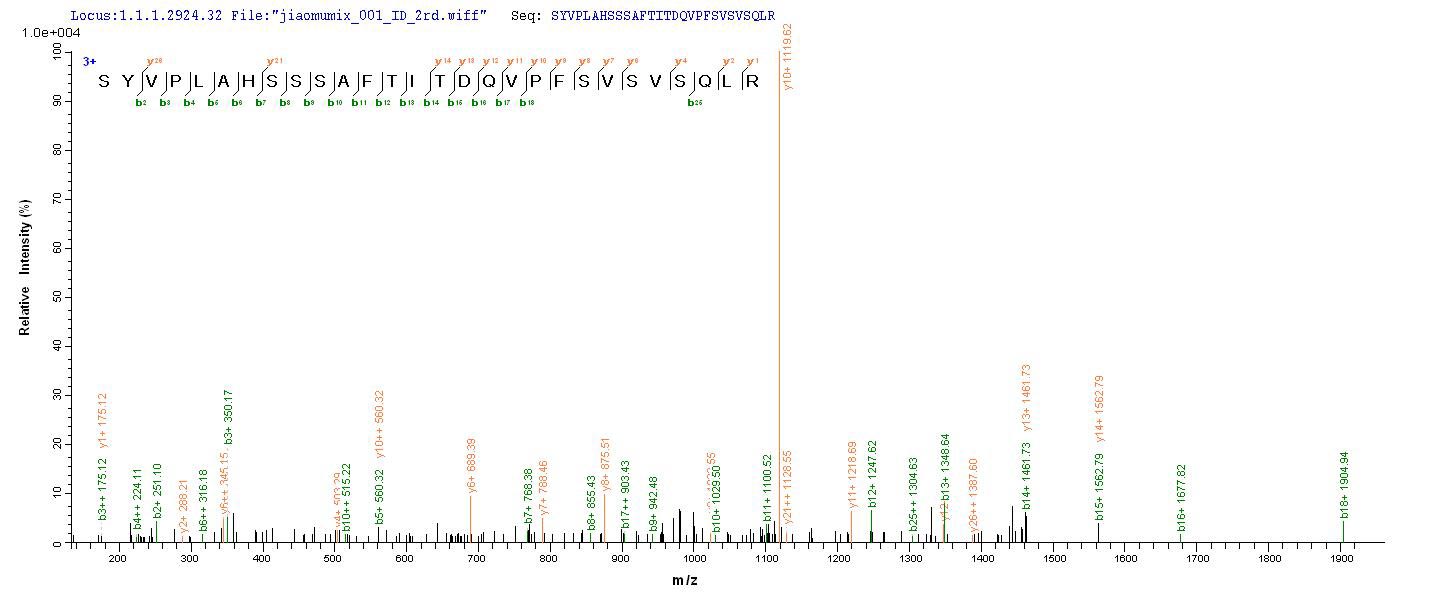
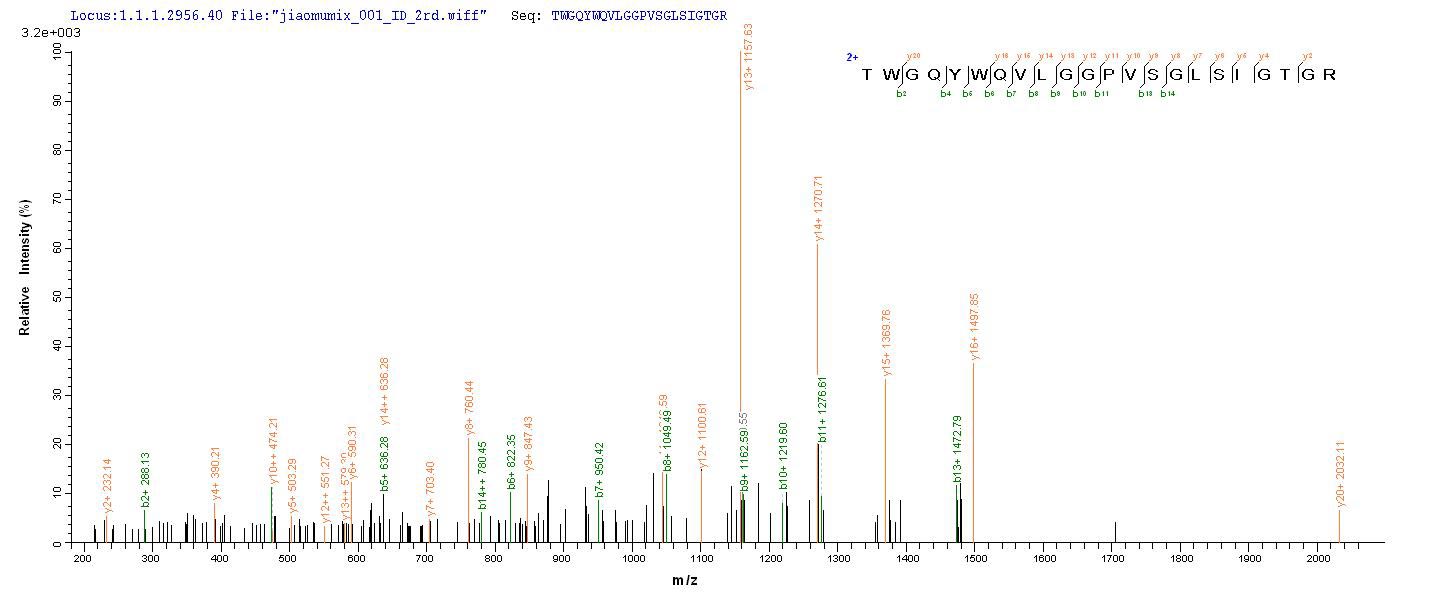
An ORF cDNA corresponding to the peptide of Human Melanocyte protein PMEL was expressed with an N-terminal 6xHis-tag in the yeast cells. PMEL CSB-YP021324HU is a truncated molecule with amino acids of Lys25-Val467. Its purity is greater than 90% assessed by SDS-PAGE. A molecular mass band of about 52 kDa was presented on the gel under reducing conditions. It was also validated by the LC-MS/MS analysis. This recombinant PMEL protein may be used to produce specific anti-PMEL antibodies or in the studies of PMEL-associated metabolism.
Premelanosome Protein (PMEL) is an amyloid protein that participates in the melanosome matrix formation through the interaction with melanin. Eumelanin deposition requires the expression of PMEL and proper formation of the melanosome matrix, both of which are not crucial for the development of pheomelanic melanosomes. As a consequence, PMEL mutations lead to eumelanin defects in cattle, chicken, and mouse. PMEL mutations are also associated with pigmentary glaucoma in humans.