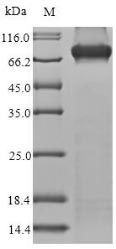
Recombinant Human Methylmalonyl-CoA mutase, mitochondrial (MMUT) is expressed in E.coli and contains the complete mature protein sequence from amino acids 33-750. The protein includes an N-terminal 6xHis-tag that makes purification and detection more straightforward. Purity appears to exceed 85% based on SDS-PAGE analysis, which should provide consistent results in most experimental settings. Endotoxin levels are kept low enough for research applications.
Methylmalonyl-CoA mutase, mitochondrial (MMUT) plays a key role in mitochondrial metabolism by converting methylmalonyl-CoA to succinyl-CoA. This reaction is essential for breaking down certain amino acids and fatty acids. The conversion step is important for energy production and general metabolism, which makes MMUT particularly valuable for researchers studying metabolic pathways and mitochondrial function.
Potential Applications
Note: The applications listed below are based on what we know about this protein's biological functions, published research, and experience from experts in the field. However, we haven't fully tested all of these applications ourselves yet. We'd recommend running some preliminary tests first to make sure they work for your specific research goals.
The human MMUT is a mitochondrial enzyme that requires precise folding, cofactor binding (adenosylcobalamin, or vitamin B12), and potential post-translational modifications for bioactivity. E. coli, as a prokaryotic system, lacks the mitochondrial chaperones and cannot provide the proper environment for the correct folding of this complex eukaryotic enzyme. The His tag may improve solubility, but cannot compensate for fundamental folding limitations. Without experimental validation (e.g., enzyme activity assays with methylmalonyl-CoA), the protein cannot be assumed to be correctly folded or bioactive. The >85% purity indicates low impurities but does not guarantee functional conformation.
1. Protein-Protein Interaction Studies Using His-Tag Pull-Down Assays
The His tag enables pull-down assays to identify binding partners, but if MMUT is misfolded, interactions may be non-physiological. The tag itself may cause artifactual binding. Validate any identified partners using native MMUT from mitochondrial preparations or confirm with co-immunoprecipitation from eukaryotic cells.
2. Antibody Development and Validation
The recombinant MMUT can serve as an immunogen for generating antibodies against linear epitopes, even if misfolded. The high purity supports consistent immunization. However, antibodies generated may not recognize conformational epitopes of native, properly folded and cofactor-bound MMUT. Validate antibody specificity against endogenous MMUT from human mitochondrial extracts.
3. Biochemical Characterization and Enzyme Kinetics Analysis
This application requires confirmed bioactivity. If MMUT is verified to be active (e.g., through methylmalonyl-CoA conversion assays), it can be used for kinetic studies and cofactor requirements. Without activity validation, data on enzyme kinetics are unreliable. First, perform a functional assay with adenosylcobalamin and substrate to confirm activity.
4. Comparative Structural and Stability Studies
Suitable for basic biophysical analysis (e.g., circular dichroism for secondary structure, thermal stability assays). However, data may not reflect native MMUT properties due to potential misfolding. The His tag may influence the results. Use the protein for stability studies, but interpret results cautiously without native MMUT controls.
Final Recommendation & Action Plan
Before using this recombinant MMUT for functional applications, validate its folding and bioactivity. Start with an enzyme activity assay using methylmalonyl-CoA as substrate and adenosylcobalamin as cofactor to confirm catalytic function. If active, proceed with interaction or kinetic studies; if inactive, limit use to non-functional applications like antibody production (with validation against native MMUT). For reliable results, consider expressing MMUT in a eukaryotic system (e.g., yeast with mitochondrial targeting) that can provide proper folding environment and cofactor incorporation. Always include appropriate controls with native MMUT when possible.