The Rat Pulmonary Surfactant-associated protein D (SP-D) ELISA kit can be your reliable and efficient solution for measuring the levels of Pulmonary surfactant-associated protein D in serum, plasma, and tissue homogenates from Rattus norvegicus (Rat) samples.
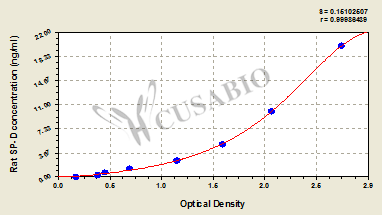
With a detection range of 0.312 ng/mL-20 ng/mL and a sensitivity of 0.078 ng/mL, this ELISA kit provides accurate and precise measurements of SP-D levels. The assay time ranges from 1-5 hours and requires a sample volume of 50-100ul, making it convenient and easy to use in your lab.
The assay principle is quantitative and the measurement method is a Sandwich assay, ensuring reliable and reproducible results. The detection wavelength is 450 nm.
Designed for research in Lung-related disease and Signal Transduction, this ELISA kit has been cited in more than 3 publications, attesting to its quality and reliability.