Proteins, the executor of life activities, efficiently execute cellular functions through interactions, frequently in the form of protein-protein interactions. Understanding any cellular process requires knowing which proteins are involved, how these proteins interact with each other, and how these interactions are controlled.
In this article, you will learn what protein-protein interactions are, how proteins interact with each other, what factors affect protein-protein interactions, protein interaction methods, and understand the association between protein-protein interactions and diseases.
Table of Contents
1. What Are Protein-Protein Interactions?
Protein-protein interactions refer to specific and intentional physical contacts established between two or more proteins to carry out specific biological activity.
Figure 1. Protein-protein interaction docking [1]
Proteins interact with their binding partners through several non-covalent interactions, including electrostatic forces, hydrogen bonds, van der Waals forces, and hydrophobic interactions. These interactions can maintain the structure and stability of proteins, promote the function of proteins, and also provide a suitable environment for various biochemical reactions in cells.
Proteins can also interact with each other through covalent bonds, chemical bonds formed by REDOX reactions. Covalent bonds allow proteins to form stable connections with other molecules for long periods, allowing for longer-lasting interactions.
In a cell or organism, a protein may interact with multiple proteins as a result of specific biochemical events, forming protein complexes. Computational techniques and statistical analysis can generate a network based on these protein-protein interactions, known as a protein-protein interaction network.
2. Why Are Protein-Protein Interactions Important in Biology?
Protein-protein interactions play a fundamental role in nearly all cellular processes, including enzymatic reactions, antibody-antigen complexes, large biomolecular assemblies, signal transduction, intercellular communication, metabolic pathways, transport, gene expression, and cell growth and proliferation.
Understanding protein-protein interactions is crucial for unraveling the complexities of biological systems, deciphering how cells work, and revealing the functions of unknown proteins. By studying these protein interactions, researchers can gain insights into the underlying mechanisms of diseases, identify potential drug targets, and develop new therapeutic strategies.
3. What Are the Factors Contributing to Protein-Protein Interactions?
In biology, both in vivo and in vitro experiments are crucial for advancing scientific understanding and addressing various research questions. Each approach offers unique advantages and serves specific purposes in studying biological systems.
4. Types of Protein-Protein Interactions
Protein-protein interactions are classified based on three criteria, including duration, composition, and binding affinity, which provide insights into the diversity, complexity, and functional significance of protein-protein interactions in biological systems, facilitating the study and understanding of protein interaction networks and their roles in cellular physiology and disease.
| Classification |
Description |
Examples |
| Based on duration |
Stable interaction |
Long-lasting associations between proteins, often involved in structural integrity or complex formation |
Haemoglobin structure, cytochrome c |
| Transient interaction |
Brief and dynamic associations between proteins, typically involved in signaling or regulatory processes |
PPIs involving muscle contraction; the transient binding of kinase and substrate during phosphorylation events |
| Based on composition |
Homo-oligomers |
Involve the binding of identical protein subunits to form oligomers |
PPIs in muscle contraction |
| Hetero oligomer |
Different protein subunits interact in hetero-oligomers; essential to control several cellular functions |
PPI between cytochrome oxidase and TRPC3; GPCRs: Gα subunit binds to the Gβγ subunit to form a heterotrimeric complex |
| Based on binding affinity |
Non-obligate |
Transient or reversible interactions that occur under specific conditions or in certain cellular contexts |
Interactions between regulatory proteins and their targets |
| Obligate |
Essential and stable interactions required for protein function or complex formation |
PPIS involved in metabolic pathways, signal transduction |
5. How to Measure Protein-Protein Interactions
Identifying protein-protein interactions is crucial for comprehending how proteins collaborate within a cell to execute various cellular functions. Experimental technologies are adept at identifying protein-protein interactions, while computational methods excel in protein-protein interaction prediction.
Assays for identifying protein-protein interactions are grouped into three categories: in vitro, in vivo, and in silico.
Table 1. Common protein interaction techniques [2]
| Approach |
Technique |
Description |
| In vitro (experimental) |
Performed in a controlled environment outside a living organism |
Tandem affinity purification-mass spectroscopy (TAP-MS) |
Based on the double tagging of the protein of interest on its chromosomal locus, followed by a two-step purification process and mass spectroscopic analysis; can identify a wide range of protein complexes and test the activeness of monomeric or multimeric protein complexes |
| Affinity chromatography |
Highly responsive, can even detect weakest interactions in proteins, and also tests all the sample proteins equally for interaction ;with coupled protein in the column |
| Co-immunoprecipitation |
A protein (bait) is immunoprecipitated with a specific antibody, its interacting protein (prey) is indirectly co-precipitated; determine whether two target proteins are bound; identify a novel role for a given protein; isolate and obtain the interacting protein complex in its natural state |
| Protein microarrays |
Microarray-based analysis allows the simultaneous analysis of thousands of parameters within a single experiment; achieve efficient and sensitive high-throughput protein analysis |
| Protein-fragment complementation ;assay (PCA) |
Used to detect PPI between proteins of any molecular weight and expressed at their endogenous levels |
| Phage display |
Originated in the incorporation of the protein and genetic components into a single phage particle |
| X-ray crystallography |
Enable visualization of protein structures at the atomic level and enhance the understanding of protein interaction and function |
| Nuclear magnetic resonance (NMR) spectroscopy |
Magnetically active nuclei oriented by a strong magnetic field absorb electromagnetic radiation at characteristic frequencies governed by their chemical environment; can even detect weak protein-protein interactions |
| In vivo ;(experimental) |
Conducted on the whole living organism itself |
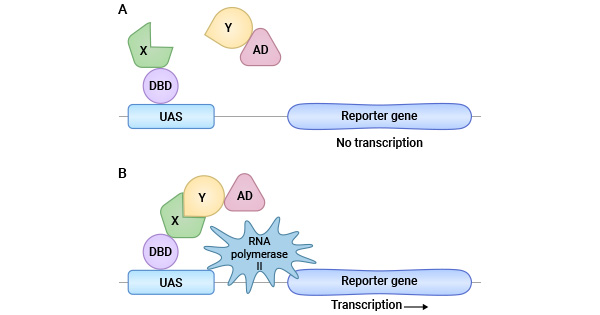
Yeast two-hybrid (Y2H) |
Typically carried out by screening a protein of interest against a random library of potential protein partners; allow the direct recognition of PPI between protein pairs; simple organization and easy detection for the transient interactions |
| Synthetic lethality |
Based on functional interactions rather than physical interaction |
| In silico ;(computational) |
Performed on a computer (or) via computer simulation |
Ortholog-based sequence approach |
Based on the homologous nature of the query protein in the annotated protein databases using pairwise local sequence algorithm |
| Domain-pairs-based sequence approach |
Predict protein interactions based on domain-domain interactions |
| Structure-based approaches |
Predict protein-protein interaction if two proteins have a similar structure (primary, secondary, or tertiary) |
| Gene neighborhood |
If the gene neighborhood is conserved across multiple genomes, then there is a potential possibility of the functional linkage among the proteins encoded by the related genes |
| Gene fusion |
Often called Rosetta stone method; ;based on the concept that some of the single-domain containing proteins in one organism can fuse to form a multidomain protein in other organisms |
| In silico ;2 hybrid (I2H) |
Based on the presumption that interacting proteins should undergo coevolution in order to keep the protein function reliable |
| Phylogenetic tree |
Predict the protein-protein interaction based on the conception that the interactive proteins show similarity in their evolution history |
| Phylogenetic profile |
Predict the interaction between two proteins if they share the same phylogenetic profile |
| Gene expression |
Predict interaction based on the conception ;that proteins translated from the genes belonging to the common expression-profiling clusters are more likely to interact with each other than proteins from the genes belonging to different clusters |
Figure 2. Yeast two-hybrid technique [3]
(Note: X: Bait protein, Y: Prey protein, DBD: DNA binding domain, AD: Activation domain, UASA: Upstream activation domain)
CUSABIO can provide phage display service for researchers to aid in investigating protein-protein interactions.
6. Protein-Protein Interactions and Disease Research
Proteins play a fundamental role in biological functions, with their interactions governing molecular and cellular mechanisms that regulate both healthy and diseased states in organisms.
Mutations disrupting the binding interface or inducing dysfunctional allosteric changes in proteins are common triggers for diseases. Therefore, exploring protein-protein interaction networks provides insights into the molecular mechanisms of diseases, offering valuable information for prevention, diagnosis, and treatment strategies.
Protein-protein interactions can contribute to disease development through various mechanisms:
6.1 Dysregulated Signaling Pathways
Aberrant protein-protein interactions can lead to abnormal activation or inhibition of signaling pathways involved in cell growth, survival, and differentiation.
In cancer, mutations in proteins involved in signaling pathways can disrupt protein-protein interactions, resulting in uncontrolled cell proliferation.
In colorectal cancer, mutations in the APC gene disrupt its interaction with β-catenin, leading to constitutive activation of the Wnt signaling pathway and uncontrolled cell proliferation [4].
Mutations in the EGFR gene can alter its interaction with downstream signaling proteins like PI3K/Akt and Ras-MAPK, promoting cell survival and proliferation in non-small cell lung cancer [5,6].
Mutations in the BRCA1 gene disrupt its interaction with other proteins involved in DNA repair and genomic instability, increasing the risk of breast and ovarian cancer in women [7].
6.2 Misfolded Proteins and Aggregation
Changes in protein-protein interactions can affect the function of proteins involved in critical cellular processes.
For instance, misfolded proteins in neurodegenerative diseases like Alzheimer's and Parkinson's can form aberrant interactions, which lead to protein aggregation, interfering with normal cellular functions and causing neuronal damage and cell death.
In Huntington's disease, mutant huntingtin protein forms abnormal interactions with various HTT-interacting proteins, leading to the formation of toxic aggregates and neuronal dysfunction [8].
In Alzheimer's disease, the interaction between amyloid-beta and tau proteins promotes the formation of neurotoxic aggregates, leading to neuronal death [9].
Mutations in the CFTR gene disrupt its interaction with chaperone proteins, causing misfolding and dysfunction of the CFTR protein in cystic fibrosis [10].
6.3 Immune System Dysfunction
Disrupted protein-protein interactions can impair immune responses, leading to susceptibility to infections or autoimmune diseases.
For instance, aberrant interactions between proteins involved in immune regulation can trigger an inappropriate immune response against self-tissues, leading to autoimmune disorders like rheumatoid arthritis and lupus.
In systemic lupus erythematosus (SLE), autoantibodies and immune complexes disrupt the interaction between regulatory T cells and dendritic cells, leading to impaired immune tolerance and autoimmune reactions [11].
In HIV/AIDS, the viral protein Vif interacts with the host APOBEC3G protein, preventing its antiviral activity and promoting viral immune evasion [12,13].
6.4 Viral Pathogenesis
Protein-protein interactions play pivotal roles in virus pathogenesis by mediating various steps of the viral life cycle, including viral entry, replication, assembly, and egress.
In influenza virus infection, viral proteins interact with host factors to hijack cellular machinery for viral replication and spread.
During HCV infection, viral NS3/4A protein inactivates the Riplet-involved RIG-I- and MAVS-independent signaling pathway, inhibiting the host antiviral response and facilitating viral replication [14].
In Zika virus infection, viral NS5 protein targets human STAT2 for proteasome-mediated degradation, inhibiting IFNAR signaling and promoting viral replication and pathogenesis [15].
In COVID-19, the interaction between the SARS-CoV-2 spike protein and the ACE2 receptor on host cells facilitates viral entry and infection [16].
In Conclusion
Protein-protein interactions reflect how proteins work together in cells to perform cellular functions in a coordinated manner. Abnormal protein-protein interactions involving endogenous proteins, proteins from pathogens or both lead to various human diseases.
Therefore, protein-protein interaction networks aid in comprehending the mechanisms behind biological responses and the pathogenic and physiological mechanisms underlying the initiation and advancement of various diseases.
Targeting protein-protein interactions is an alternative option in developing targeted therapies that can intervene at the molecular level, providing a critical strategy for the development of novel drugs.
Further Considerations
In addition to protein-protein interactions, there are other types of interaction, such as protein-DNA interaction and protein-RNA interaction.
Protein-DNA interactions involve proteins binding to specific sequences on DNA molecules, which can influence the function and activity of both the protein and the DNA. They play an important role in many biological processes, including gene regulation, DNA replication, repair, and recombination.
Protein-RNA interactions involve RNA-binding proteins (RBPs) binding to RNA sequences or structural elements, which occur at different stages of RNA metabolism and can influence the fate and function of RNA molecules. They play essential roles in gene expression, RNA processing, localization, stability, and translation.
References
[1] Stanzione, F., Giangreco, I., & Cole, J. C. (2020). Use of molecular docking computational tools in drug discovery [J]. Progress in Medicinal Chemistry, 60, 273-343.
[2] Rao VS, Srinivas K, Sujini GN, Kumar GN. Protein-protein interaction detection: methods and analysis [J]. Int J Proteomics. 2014;2014:147648.
[3] Farooq, A., Shaukat, Z., Aiman, S., & Li, H. (2021). Protein-protein interactions: Methods, databases, and applications in virus-host study [J]. World Journal of Virology, 10(6), 288-300.
[4] Hankey W, Frankel WL, Groden J. Functions of the APC tumor suppressor protein dependent and independent of canonical WNT signaling: implications for therapeutic targeting [J]. Cancer Metastasis Rev. 2018 Mar;37(1):159-172.
[5] Liang, S. I. et al. Phosphorylated EGFR dimers are not sufficient to activate ras [J]. Cell Rep. 22, 2593–2600 (2018).
[6] Iyer, R.S., Needham, S.R., Galdadas, I. et al. Drug-resistant EGFR mutations promote lung cancer by stabilizing interfaces in ligand-free kinase-active EGFR oligomers [J]. Nat Commun 15, 2130 (2024).
[7] Savage, K. I., & Harkin, D. P. (2015). BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability [J]. The FEBS Journal, 282(4), 630-646.
[8] Liu, L., Tong, H., et al. (2022). Huntingtin Interacting Proteins and Pathological Implications [J]. International Journal of Molecular Sciences, 24(17), 13060.
[9] Gallardo, G.; Holtzman, D.M. Amyloid-β and Tau at the Crossroads of Alzheimer’s Disease [J]. Adv. Exp. Med. Biol. 2019, 1184, 187–203.
[10] Lim, S. H., Legere, A., Snider, J., & Stagljar, I. (2017). Recent Progress in CFTR Interactome Mapping and Its Importance for Cystic Fibrosis [J]. Frontiers in Pharmacology, 8.
[11] Justiz Vaillant AA, Goyal A, Varacallo M. Treasure Island: StatPearls Publishing; 2022. Systemic Lupus Erythematosus.
[12] Stopak, K., de Noronha, C., et al. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability [J]. Mol. Cell 12, 591–601 (2003).
[13] Li, Y., Langley, C. A., et al. (2023). The structural basis for HIV-1 Vif antagonism of human APOBEC3G [J]. Nature, 615(7953), 728-733.
[14] Vazquez, C., Tan, C. Y., & Horner, S. M. (2019). Hepatitis C Virus Infection Is Inhibited by a Noncanonical Antiviral Signaling Pathway Targeted by NS3-NS4A [J]. Journal of Virology, 93(23).
[15] Best, S. M. (2017). The Many Faces of the Flavivirus NS5 Protein in Antagonism of Type I Interferon Signaling [J]. Journal of Virology, 91(3).
[16] Zhou, Y., Liu, Y., Gupta, S. et al. A comprehensive SARS-CoV-2–human protein–protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets [J]. Nat Biotechnol 41, 128–139 (2023).
CUSABIO team. Exploring Protein-Protein Interactions: A Comprehensive Guide to Understanding and Utilizing It. https://www.cusabio.com/c-21172.html




Comments
Leave a Comment