Call us
301-363-4651 (Available 9 a.m. to 5 p.m. CST from Monday to Friday)
| Code | CSB-EL33244 |
| Size | 96T,5×96T,10×96T |
| Price | Request a Quote |
| Trial Size |
24T ELISA Kit Trial Size (Only USD$150/ kit) * Sample kit cost can be deducted as a $30 credit for each 96-assay kit of the same analyte and brand you subsequently purchase within six months until depleted. More details >> Interested in a trial size? Please leave a message below.
|
| Have Questions? | Leave a Message or Start an on-line Chat |
| Intra-assay Precision (Precision within an assay): CV%<8% | ||
| Three samples of known concentration were tested twenty times on one plate to assess. | ||
| Inter-assay Precision (Precision between assays): CV%<10% | ||
| Three samples of known concentration were tested in twenty assays to assess. | ||
| Intra-Assay Precision | Inter-Assay Precision | |||||
| Sample | 1 | 2 | 3 | 1 | 2 | 3 |
| n | 20 | 20 | 20 | 20 | 20 | 20 |
| Mean(ng/ml) | 10.79 | 11.06 | 11.36 | 10.85 | 10.99 | 10.02 |
| SD | 0.032 | 0.034 | 0.039 | 0.041 | 0.045 | 0.041 |
| CV(%) | 5.53 | 5.74 | 6.43 | 7.05 | 7.64 | 7.55 |
These standard curves are provided for demonstration only. A standard curve should be generated for each set of samples assayed.
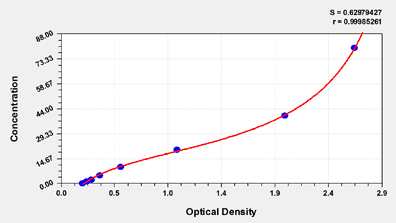
| ng/ml | OD1 | OD2 | Average | Corrected |
| 80 | 2.648 | 2.594 | 2.621 | 2.409 |
| 40 | 1.973 | 2.032 | 2.003 | 1.791 |
| 20 | 1.027 | 1.069 | 1.048 | 0.836 |
| 10 | 0.559 | 0.537 | 0.548 | 0.336 |
| 5 | 0.365 | 0.358 | 0.362 | 0.150 |
| 2.5 | 0.291 | 0.285 | 0.288 | 0.076 |
| 1.25 | 0.251 | 0.240 | 0.246 | 0.034 |
| 0 | 0.213 | 0.211 | 0.212 |
There are currently no reviews for this product.