1. What Is Phage Display
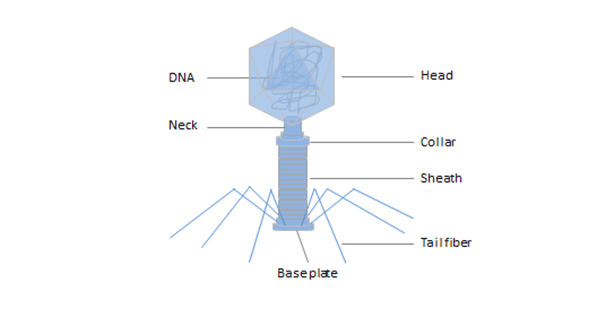
Bacteriophage (phage) is a virus that infects and replicates within bacteria and archaea, for following the injection of their genome into its cytoplasm. Phage is composed of proteins that encapsulate a DNA or RNA genome, and may have relatively simple or elaborate structures.
Phage display was first developed by G. Smith in 1985[1] as a method of presenting polypeptides on the surface of lysogenic filamentous bacteriophages. In phage display technique, a gene encoding a protein of interest is inserted into a phage coat protein gene, causing the phage to display the protein on the outside. And containing the gene for the protein inside, resulting in a connection between genotype and phenotype.
These displaying phages can be screened for other proteins, peptides or DNA sequences, in order to detect interaction between the displayed protein and those other molecules. So, it has become one of the most powerful and widely used laboratory technique for the study of protein-protein, protein-peptide and protein-DNA interactions.
In this way, large libraries of proteins can be screened and amplified in the process called in vitro selection, which is analogous to natural selection. Phage display is also an effective way for producing large amounts of peptides, proteins and antibodies.
Phage display technology has evolved into an extremely versatile and powerful platform for protein engineering.
2. Phage Display Protocol
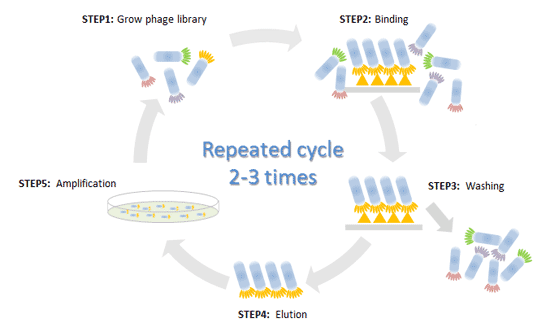
In general, there are 5 steps for phage display technology as below:
STEP1: Construct phage display library
Recombinant DNA technology is used to incorporate foreign cDNA into viral DNA. Different sets of genes are inserted into the genomes of multiple phages. Spliced into gene for a coat protein, so that the protein will be displayed on the outside of phage particles, and these separate phages will only display one protein, peptide, or antibody.
Collections of these phages can comprise libraries, such as antibody phage library, protein phage library, or random phage library.
STEP2: Binding
These libraries are exposed to selected targets and only some phages will interact with targets. The target is for which specific ligands planned to be identified such as immobilized protein, cell surface protein or vascular endothelium.
STEP3: Washing
Unbound phages can be washed away, and only those which showing affinity for the receptors was left.
STEP4: Elution
Recovery of the target bound phage by elution.
STEP5: Amplification
Eluted phages showing specificity are used to infect new host cells for amplification, or direct bacterial infection and amplification of the recovered phage.
Back to step 1, repeated cycle 2-3 times for stepwise selection of best binding sequence. After that, you can Enrichment and purification the phage repertoire by precipitation methods to increasing the phage titer.
Phage display is the technology that looks simple but hard to do. With years of experience, we can offer phage display service such as high-quality phage display library construction and custom phage display library screening services to meet your various demands precisely.
3. Classification of Phage Display Systems
The most common bacteriophages used in phage display are E.coli filamentous bacteriophages (f1, fd, M13) [2] , though T4[3], T7, and λ phage have also been used.
3.1 E.coli filamentous bacteriophages (f1, fd, M13)
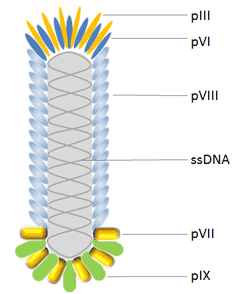
A filamentous bacteriophage is a type of phage, defined by its filament-like or rod-like shape. Filamentous phages usually contain a genome of single-stranded DNA and infect Gram-negative bacteria. The family of Ff, M13, fd, and f1 are vital phages which have utility in phage display among which M13 phage is the most generally used[4].
M13 bacteriophage has a cylindrical shape with a length of 880nm and a diameter of 6nm. It encapsulates a single-strand genome that encodes five different capsid proteins which comprise two groups, major coat proteins (pVIII) and minor coat proteins (pVII, pIX, pVI and pIII).
E. coli filamentous bacteriophages are commonly used for phage display. Most antibodies and peptides are displayed at phage proteins pIII[5] and pVIII[6], which constructed pIII and pVIII display system. Moreover, hybrid phage system enables displaying large proteins with all five M13 coat proteins as N-terminal fusions with pIII, pVIII, pVII and pIX, and also as C-terminal fusions with pVI, pIII, and pVIII.
pIII is the protein that determines the infectivity of the virion. It consists of 406 amino acid residues and occurs at the phage tip in 3 to 5 copies[7]. An advantage of using pIII rather than pVIII is that pIII allows for monovalent display when using a phagemid combined with a helper phage. Moreover, pIII allows for the insertion of larger protein sequences (>100 amino acids)[8] and is more tolerant to it than pVIII.
pVIII is the main coat protein of Ff phages, which is expressed by gene 8 and occurs in 2700 copies. Therefore it is used to enhance detection signal when phage displayed antibody associates with antigen. Peptides are usually fused to the N-terminus of Pviii which are usually 6-8 amino acids long[7] . This makes the use of this protein unfavorable for the discovery of high affinity binding partners. Moreover modifications of pVIII are made to increase the efficiency of display onto pVIII[9], and now there has been great progress.
pVI has been widely used for the display of cDNA libraries, which is an attractive alternative to the yeast-2-hybrid method for the discovery of interacting proteins and peptides due to its high throughput capability[7]. pVI has been used preferentially to pVIII and pIII for the expression of cDNA libraries because one can add the protein of interest in the C-terminus of pVI without greatly affecting pVI's role in phage assembly.
pVII and pIX, located to the phage tip opposite that of pIII, may both complement current phage display systems and be used as alternative scaffolds for display and selection to further improve phage display as the ultimate combinatorial engineering platform.
3.2 T4 Phage
T4 phage (Enterobacteria phage T4) is a bacteriophage that infects E.coli bacteria. It is a member of the T-even phages, a group including enterobacteriophages T2 and T6. T4 phage is a relatively large phage, at approximately 90 nm wide and 200 nm long. It’s double-stranded DNA genome is about 169 kbps long[10] and encodes 289 proteins. T4 phage is built with three essential proteins: gp23, which forms the hexagonal capsid lattice; gp24, which forms pentamers at eleven of the twelve vertices; and gp20, which forms the unique dodecameric portal vertex through which DNA enters during packaging and exits during infection[11].
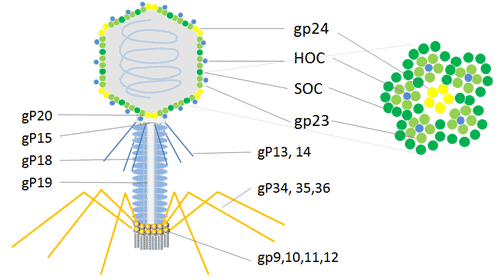
In addition to the essential capsid proteins, gp23, gp24, and gp20, the T4 capsid is decorated with two nonessential outer capsid proteins: HOC (highly antigenic outer capsid protein) and SOC (small outer capsid protein). Both HOC and SOC are dispensable, and bind to the capsid after the completion of capsid assembly[11]. Null (amber or deletion) mutations in either or both the genes do not affect phage production, viability, or infectivity.
HOC and SOC are dispensable T4 capsid proteins that can be used for phage display of multiple copies of peptides and proteins. The phage T4 HOC, SOC bipartite display system is attractive for the expression of cDNA and display of peptides or proteins at high copy numbers on the phage capsid surface[12]. It could be applied to cDNA expression, displays larger proteins in high copy number and inserts into stop codon on the C-terminal of SOC protein that occurs in 810 copies or N-terminal of HOC protein that occurs in 155 copies[13].Therefore, the phage T4 dual site display emerges as a powerful method with an enhanced immune response in animals for research and development of immunological products.
3.3 T7 Phage
T7 phage is an icosahedral virus of the Podoviridae family and has a linear double stranded (ds) DNA genome. Similar to T4, bacteriophage T7 possesses a head and tail structure. The icosahedral head, where T7 conserves its dsDNA genome, is composed of 415 copies of capsid gp10, arranged as 60 hexamers on the surface and 11 pentamers at the vertices. There exist two isoforms of major coat protein gp10, gp10A and gp10B, in a nine-to-one ratio, resulted from natural translational frame shift at amino acid 341.
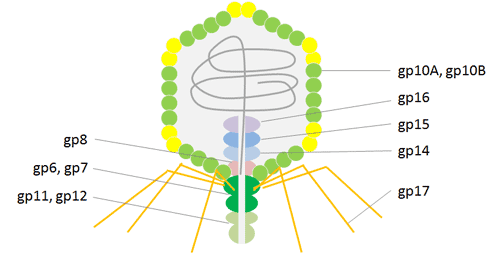
The minor protein gp10B results from a frame shift in the end of the gene that makes the capsid protein 52 residues longer[14]. Fusion proteins are displayed on protein gp10B C-terminally of the 52 extra residues. So that it can avoid problems associated with steric hindrance. T7 phage particles exhibit high stability under various extreme conditions, including high temperature and low pH, which facilitates effective high throughput affinity elutriation. Recent applications of the T7 phage display system have been instrumental in uncovering mechanisms of molecular interaction, particularly in the fields of antigen discovery, vaccine development, protein interaction, and cancer diagnosis and treatment.
3.4 lambda phage
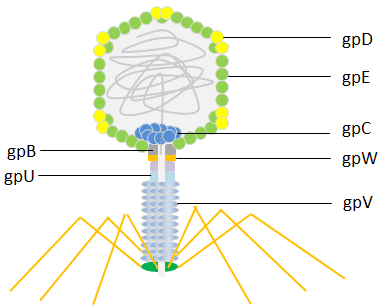
lambda phage (Enterobacteria phage λ, coliphage λ) is a bacterial virus which infects the bacterial species E. coli. It was discovered by Esther Lederberg in 1950[15]. The lambda phage has an icosahedral head. The main structure of the shell is built from the major coat protein gpE (415 copies) and is stabilized by the capsid protein gpD (402–420 copies) [16]. The head is linked to a flexible helical tail constructed by disks of the major tail protein gpV.
Both the tail protein gpV and the head protein gpD have been used for phage display. Initially, the lambda foo vector was constructed for the C-terminal display on gpV, with a low display level that made it suitable for capturing high-affinity interactions [17]. Later, systems were developed for the display of peptides N-terminally or C-terminally to the major coat protein gpD [18]. The lambda phage has been engineered to display efficiently multiple copies of peptides or even large protein domains providing a powerful tool for screening libraries of peptides, proteins and cDNA.
4. Applications of Phage Display
Nowadays, phage display as a rapidly developing technology has been used in a wide range of applications in different research areas including epitope mapping, identification of new receptors & ligands, in vitro protein evolution, drug discovery, and so on. Some of the most successful applications of phage display are explained in the following sections.
4.1 Epitope mapping and mimicking
Upon encountering antigen, host humoral immunity activates and triggers production of antibodies which directed against foreign protein epitopes. Knowledge of these protein epitopes is pivotal in understanding the pathogenesis of pathogen infections and in developing diagnostic reagents, therapeutic antibodies, and effective vaccines.
Peptide phage display libraries are useful tools for identification of continuous or linear epitopes involving in interaction with antibody. Isolation and identification of mimotopes from peptide phage libraries is powerful approach to improve immunological studies in order to design and develop vaccine candidate [19].
4.2 Identification of new receptors & ligands
Phage display method using either gene-specific libraries, or random peptide libraries, provides a powerful technique for an approach to epitope identification[20]. The technique can identify amino acids on protein antigens that are critical of antibody binding.
The random peptide sequence was displayed on the surface of the phage to obtain the phage display polypeptide library. Polypeptides identifying specific cells were obtained by differential screening using cells as screening targets. By studying the polypeptide sequence, we can further to obtain the receptor protein expressed specifically on the cell surface.
4.3 Protein-protein interaction studies
Protein–protein interactions mediate essentially all biological processes. A detailed understanding of these interactions is thus a major goal of modern biological chemistry. Phage display is a potent and versatile method for studying protein-protein interaction[21]. It can be applied to a wide range of protein interaction partners and used in a number of applications, especially in mapping intracellular interactions of the distinct protein domain.
The polypeptide library displayed by bacteriophages is composed of random short peptide sequences of specific length. Short peptide sequences can be obtained by affinity screening for the random library by target proteins (such as receptors). Sequence analysis and synthesis of corresponding short peptides, and then we can study the interaction between two proteins.
Recombinant antibodies are useful tools for therapy, diagnosis and research. With major developments in molecular biology, numerous display technologies have been successfully introduced for recombinant antibody production. Even so, antibody phage display still remains the gold standard for recombinant antibody production. Its success is mainly attributed to the robust nature of phage particles allowing for automation and adaptation to modifications.
a. Production of gene fragment: This phase involves animal immunization with the desired antigen and then isolation of B lymphocytes, mRNA extraction and cDNA synthesis. The synthesized cDNA contain genetic information of all antibodies targeting various antigens and consist of approximately 109 to 1011 lymphocyte clones.
b. Cloning of gene fragments in the phagemid vectors: Genes related to the different clones of antibodies are digested with restriction enzymes, clone into phagemid vectors and then display on the surface of phages. Sequence diversity of the fragments at this step leads to optimum isolation of antibodies in the later steps. The phagemid vectors need helper phage to package and exit from the bacterial cells and enter to the medium.
c. Selection of specific phages: After cloning of the fragments into phagemid vectors, variety of clones of antibodies display on the surface of the phage. Selections of specific clone that recognize the antigen (target of interest) perform by biopanning. Then the phage carrying specific antibody can be isolated from non-specific phages due to antigen–antibody binding properties.
d. Screening: Isolation of antibodies with high affinity to target is the main aim of this step. Screening is performed using different methods like: immunoassay, immunocytochemistry, active isolation of cells due to their fluorescent properties and immunoblotting.
You can construction phage display library by yourself or choose some phages display companies to do it. Also you can by a pre-made library directly. If you already had a pre-made library, you can just start at selection. Pre-made antibody libraries represent a naive antibody repertoire which consists billons of different antibody scFv or fad genes. You should select the ones that actually recognize your antigen. If the repertoire is diverse enough, there are antibodies against your antigen present, and you just have to enrich them.
4.5 Protein directed evolution
Phage display technology as a selection based system is an attractive method for evolution of new biological drugs. Directional transformation protein refers to use cassette mutation, error-prone PCR method to mutant protein or a particular cod sequence structure domain. Proteins or structural domain mutations present library will be display in phage surface. Then we can obtain the required have directional change of phage clones by affinity screening. The primary structure can be derived from the sequence of DNA that can be used to screen more cytokines receptors ability, new enzyme inhibitors, DNA transcription factors in combination with the new sites, new cytokine antagonists, new enzymes and enhance biological active protein.
4.6 Drug discovery
Peptides as biologically active molecules in hormones, neurotransmitters, cytokines, antigens, and growth factors are involved in a wide variety of biological processes. So, peptides are extensively used as therapeutic and diagnostic agents in the medical fields such as oncology, endocrinology, urology, and obstetrics. The peptide phage libraries with presenting a huge number of different peptides mimicking the genuine epitopes play a key role in the development of new therapeutic peptides. Until now, several peptide drugs have been developed using phage display technology[22].
4.7 In vitro diagnostic
At present, phage display library technology is broadly employed to examine host-pathogen interaction, development of disease diagnostic markers, and identification of vaccine candidates and novel antibodies against pathogenic targets.
The cell-surface antigens in pathogens as molecular binding sites are suitable targets for vaccine development, since these antigens can affect the bacterial division, replication, and virulence of pathogens. There are two distinct strategies for application of phage display technology in infectious diseases area: In the pathogen-targeted phage display, molecular targets such as cell replication enzymes or host-pathogen virulence factors are targeted for screening; while in the cell-based screening method, bacterial whole cell is employed as target for screening. The cell-based screening with live pathogens can target the native cell surface proteins. This screening method is suitable when target antigen is unstable under immobilization conditions.
5. Phage display FAQ
Q1: Why can't I find a blue plaque after second round of phage amplification, with a plaque titer assay?
A1: There could be several reasons as below:
a. Please confirm if the dilution ratio is correct.
b. There are other phage contaminations in the amplification process, and the bacteria are lysed.
c. The phage has not been centrifuged when the precipitation is insufficient.
d. The x-gal is failure.
Q2: While assaying the titer of phage, why is the plate all blue?
A2: May be the titer of phage is too high, you should dilute more gradients. And please control the incubation time, so that the bacterial plaques not been too large to confluent.
Q3: Antibody Selection from phage display Libraries: after panning and dilution, why is there no plaque on the plate?
A3: There could be several reasons as below:
a. The medium is not suitable, may be you have added wrong antibiotic.
b. The dilution ratio of phage is too large. In general, after panning, the eluted phage titer should be 105-7.
c. The phage is not been neutralized after elution, so that it is inactivation.
d. The incubation temperature is not suitable, so that the flagella of bacteria have not been expressed.
e. Please confirm that if the host bacteria are correct, so that it can be infected, and express flagella.
Q4:How can I manipulation the plaque size of bacteriophage?
A4: The size of the plaque is a trait of the replication cycle. If the bacterial plaques are becoming confluent, you could try to do something to decrease the rate at which they grow. Such as follow:
a. Removing nutrients, you may reduce the burst size or reduce the number of subsequent infections, and subsequently produce smaller plaques.
b. Reduce temperature, or shorter incubation times.
c. Reduce the multiplicity of infection.
d. Try adding more quantity and higher concentration of host cells.
Q5: After screening for several rounds, why did I obtain the positive clones with the same sequences?
A5: There could be several reasons as below:
a. The capacity of your antibody library is too small and the diversity is poor. You can try to screening with mix multiple libraries.
b. The washing condition is too strict. You can try to reduce the concentration of detergent in the washing buffer appropriately.
c. The washing time is too excessive, results in the loss of most of the clones. You can try to reduce washing times appropriately.
d. The panning round is excessive, and you can select and check the clone after the previous screening.
e. Please determine that if the phage has a droplet of 100 times its capacity.
Q6: After screening, why are there no positive clones obtained by ELISA assay?
A6: There could be several reasons as below:
a. During the screening process, the non-specific conjugated phages are excessive, and the phages adsorbed by antigen were few. So that there are no positive clones could be selected.
b. The antibody has not been expressed, then we could determine by western blot directly.
c. The step of ELISA assay is not correct, then we could determine by positive control.
d. The single-plaque plate has been contaminated, and the E.coli with same resistant has been selected.
Q7: Why can't the screened antibodies recognize endogenous proteins?
A7: There could be several reasons as below:
a. The antibodies have been screened are not specific for target proteins, but specific for the vector or label sequence.
b. The expression of endogenous protein is too low to be detected.
c. Low antibody affinity.
References
[1] Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science, 1985, 228 (4705): 1315–7.
[2] Kehoe JW, Kay BK. Filamentous phage display in the new millennium. Chem. Rev. 2005, 105 (11): 4056–72.
[3] Malys N, Chang DY, Baumann RG, et al. A bipartite bacteriophage T4 SOC and HOC randomized peptide display library: detection and analysis of phage T4 terminase (gp17) and late sigma factor (gp55) interaction. J Mol Biol. 2002, 319 (2): 289–304.
[4] J. Rakonjac. Filamentous bacteriophages: biology and applications eLS. John Wiley & Sons Ltd, Chichester. 2012
[5] Cho W, Fowler JD, Furst EM. Targeted binding of the M13 bacteriophage to thiamethoxam organic crystals. Langmuir 2012; 28:6013-20.
[6] Hess GT, Cragnolini JJ, Popp MW, et al. M13 bacteriophage display framework that allows sortase-mediated modification of surface-accessible phage proteins. Bioconjug Chem 2012, 23:1478-87.
[7] Lowman HB, Clackson T. "1.3". Phage display: a practical approach. Oxford: Oxford University Press.2004, 10–11.
[8] Sidhu SS, Weiss GA, Wells JA. High copy display of large proteins on phage for functional selections. J. Mol. Biol.2000, 296 (2): 487–95.
[9] Hess GT, Cragnolini JJ, Popp MW, et al. M13 bacteriophage display framework that allows sortase-mediated modification of surface-accessible phage proteins. Bioconjug Chem 2012; 23:1478-87.
[10] Miller, ES, Kutter, E, Mosig, G. et al. Bacteriophage T4 genome[J]. Microbiology and molecular biology reviews : MMBR. 2003, 67 (1): 86–156.
[11] Venigalla B Rao, Lindsay W Black. Structure and assembly of bacteriophage T4 head. Rao and Black Virology Journal 2010, 7:356
[12] Malys N, Chang DY, Baumann RG, et al. "A bipartite bacteriophage T4 SOC and HOC randomized peptide display library: detection and analysis of phage T4 terminase (gp17) and late sigma factor (gp55) interaction". J Mol Biol. 2002, 319 (2): 289–304.
[13] Wu J, Tu C, Yu X, Zhang M, et al. Bacteriophage T4 nanoparticle capsid surface SOC and HOC bipartite display with enhanced classical swine fever virus immunogenicity: a powerful immunological approach. J Virol Methods. 2007;139:50–60.
[14] Danner S, Belasco JG. T7 phage display: a novel genetic selection system for cloning RNA-binding proteins from cDNA libraries. Proc. Natl. Acad. Sci. U.S.A. 2001, 98 (23): 12954–9.
[15] Esther Lederberg, Lysogenicity in Eescherichia coli strain K-12, Microbial Genetics Bulletin, 1950, v.1, pp. 5–8.
[16] Yang F, Forrer P, Dauter Z, et al. Novel fold and capsid-binding properties of the λ-phage display platform protein gpD. Nature Structural Biology. 2000;7(3):230–237.
[17] Maruyama IN, Maruyama HI, Brenner S. λfoo: a λ phage vector for the expression of foreign proteins. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(17):8273–8277.
[18] Mikawa YG, Maruyama IN, Brenner S. Surface display of proteins on bacteriophage λ heads. Journal of Molecular Biology. 1996;262(1):21–30.
[19] L.F. Wang, M. Yu. Epitope identification and discovery using phage display libraries: applications in vaccine development and diagnostics. Curr Drug Targ, 2004, 1-15.
[20] Rowley MJ1, O'Connor K, Wijeyewickrema L. Phage display for epitope determination: a paradigm for identifying receptor-ligand interactions. Biotechnol Annu Rev. 2004;10:151-88.
[21] Sachdev S. Sidhu Dr, et al. Exploring Protein–Protein Interactions with Phage Display
[22] Leila Rahbarnia, Safar Farajnia, Hossein Babaei, Jafar Majidi, Kamal Veisi, Vahideh Ahmadzadeh & Bahman Akbari (2016): Evolution of Phage Display technology: From Discovery to Application, J Drug Target. 2017 Mar;25(3):216-224
CUSABIO team. The Overview of Phage Display: Protocol, Classification, Application and FAQs. https://www.cusabio.com/c-20680.html





Comments
Leave a Comment