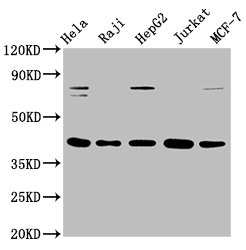
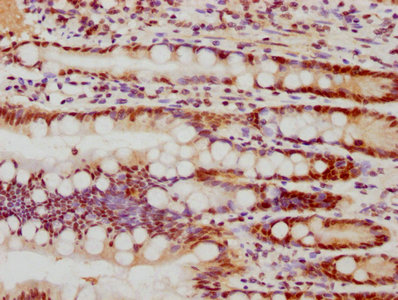
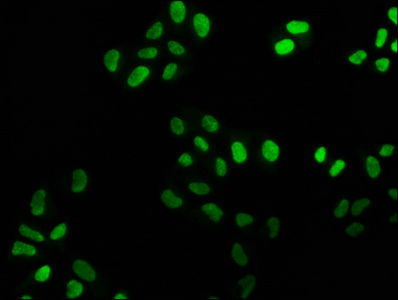
The creation of the FEN1 recombinant monoclonal antibody involves the utilization of DNA recombinant technology and in vitro genetic manipulation. The process begins by immunizing animals with a synthesized peptide derived from human FEN1, which stimulates an immune response and facilitates the isolation of B cells. Through meticulous screening and identification, positive B cells are selected for further analysis. The light and heavy chains of the FEN1 antibody are amplified using PCR and integrated into a plasmid vector. This recombinant vector is then introduced into host cells, where it drives the expression of the antibody. The FEN1 recombinant monoclonal antibody is subsequently purified from the cell culture supernatant using affinity chromatography. This highly specific antibody exhibits reactivity towards human FEN1 protein and is particularly suitable for ELISA, WB, IHC, and IF applications.
FEN1 is involved in DNA replication and repair processes in cells. FEN1 is responsible for cleaving the 5'-overhanging flap structures that arise during Okazaki fragment processing in DNA replication. FEN1 can also excise single-stranded DNA regions that have base damage or other abnormalities. In addition to its endonuclease activity, FEN1 also possesses exonuclease activity and can remove nucleotides from the 5' or 3' ends of DNA strands. FEN1's critical role in DNA repair and replication processes makes it an important protein for maintaining genome stability and preventing mutations.