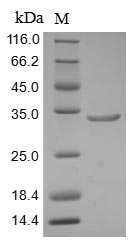
The recombinant Human ARID1A was expressed with the amino acid range of 1976-2231. The theoretical molecular weight of the ARID1A protein is 32.4 kDa. The ARID1A protein was expressed in baculovirus. The N-terminal 10xHis tag and C-terminal Myc tag was fused into the coding gene segment of ARID1A, making it easier to detect and purify the ARID1A recombinant protein in the later stages of expression and purification.
AT-rich interactive domain-containing protein 1A (ARID1A) is a highly scrutinized protein with research primarily focused on two main areas. Firstly, there is significant attention on the role of ARID1A in cancer occurrence and progression, especially concerning mutations and inactivation across various cancer types. Secondly, ARID1A's functions in genome stability and chromatin remodeling have sparked broad interest, directly implicating tumor development mechanisms. Furthermore, research on ARID1A extends to its involvement in cell cycle regulation, DNA repair, and transcriptional control. Overall, studies on ARID1A have profound implications in unraveling its contributions to cancer biology and cellular regulation.