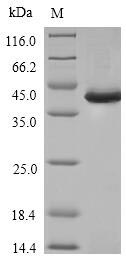
To make this Recombinant Human SPI1 protein, the SPI1 gene was isolated at first and cloned into an expression vector. CUSABIO has built a mature recombinant protein platform. This Recombinant Human SPI1 protein was developed in the platform. It was expressed in E.coli at the region of 1-270aa of the Human SPI1 protein. N-terminal 6xHis tag was fused with the expression vector for affinity and purification purposes. The purity is 85%+ determined by SDS-PAGE.
SPI1 gene encodes the hematopoietic master TF PU.1. Previous studies define the clinical and molecular phenotype of this novel inherited PU.1 haploinsufficiency syndrome, which they term PU.1-mutated agammaglobulinemia (PU.MA). PU.1 is a key transcriptional regulator required in the development of multiple hematopoietic lineages. Somatic mutations in SPI1 have been reported in the context of acute myeloid leukemia. However germline variants impacting human SPI1 have not been previously identified. Mouse models of Spi1-deficient hematopoiesis indicate a crucial role for PU.1 in early lineage commitment, differentiation of multiple myeloid lineages, as well as B cell development. However, the precise effect of SPI1 loss on human hematopoiesis has remained unknown. PU.1 has been shown to have multifaceted roles in hematopoiesis, with reduced PU.1 levels causing B cell developmental arrest, but other specific PU.1 perturbations can favor B cell over myeloid lineage development.