Universal Protein Resource (UniProt) comprises the Swiss institute of bioinformatics (SIB), the European institute of bioinformatics (EBI) and the protein information resource (PIR). Its main objective is to provide the scientific community with a central resource for protein sequence and functional information.
1. Swiss-Prot and TrEMBL
SWISS-PROT is a protein sequence database containing detailed annotations. It was established in 1986 and jointly maintained by the department of medical biochemistry of the University of Geneva and the EMBL data library (now EBI) since 1987. The database is currently merged into the UniProt database.
UniProt knowledgebase (UniProtKB) is a comprehensive knowledge base of protein sequences, which consists of two parts: UniProtKB/ SWISS-PROT and UniProtKB/TrEMBL.
1.1 Reviewed (Swiss-Prot) - Manually Annotated
All sequence entries in the SWISS-PROT database are carefully verified by experienced molecular biologists and protein chemists through computer tools and related literature.
1.2 Unreviewed (TrEMBL) - Computationally Analyzed
TrEMBL is a computer-annotated protein sequence database, which is an auxiliary database of SWISS-PROT protein sequence database. This database contains the translation content of all coding sequences (CDS) in EMBL nucleic acid sequence database which has not been incorporated into SWISS-PROT database.
The general structure and retrieval mode of SWISS-PROT and TrEMBL records is consistent. The main difference is that TrEMBL data is "Preliminary", but SWISS-PROT data is "standard".
2. Basic Situation of the SWISS-PROT
By the end of 2001, the database contained a total of 102,708 sequence data, including 37,803,202 amino acids.
Regarding the literature, the existing sequence data of the SWISS-PROT database involves 92,845 articles in 1,202 journals. There are 91 journals cited in more than 100 literatures, of which the top 20 journals frequently cited are cited more than 1,000 times. These journals are the main source of literature for publishing protein sequence related information.
In 2001, SWISS-PROT group started the Plant Proteome Annotation Program [1] [2] [3]. The goal of the project is to annotate plant-specific and plant family proteins according to the SWISS-PROT standard [4]. The current focus of the project is to manually annotate the proteome of dicotyledons (Arabidopsis thaliana) and monocotyledons (Oryza sativa).
3. Features of the SWISS-PROT
Compared with other protein databases, SWISS-PROT database differs from other protein sequence databases in three different standards:
- Annotation. SWISS-PROT database contains the protein sequences that have been carefully examined and accurately annotated in the EMBL nucleic acid sequence database.
- Minimum redundancy. Redundant sequences are minimized in SWISS-PROT.
- Integration with other databases. SWISS-PROT has cross-referenced with more than 30 other data, including nucleic acid sequence libraries, protein sequence libraries and protein structure libraries.
3.1 Annotation
SWISS-PROT provides detailed annotation information on protein sequences. Annotation include information on protein function, post-translational modification of proteins, domains and binding sites, secondary structures, quaternary structures, and diseases associated with protein deficiency.
3.2 Minimum Redundancy
In SWISS-PROT, each entry contains as much relevant literature information as possible, and is integrated. If there is disagreement, it is indicated in the Feature Table.
3.3 Integrate with Other Databases
SWISS-PROT establishes cross-references with a variety of databases databases, such as the protein tertiary structure library PDB, the human gene Mendelian genetic database (MIM), the protein type and the site library (PROSITE), which can directly access related entries in other databases. This extensive and practical database network connection allows users to obtain all aspects of protein information at the same time.
4. How to Use SWISS-PROT Database?
1. Direct access from the UniProt homepage (http://www.uniprot).
2. There are multiple sites on the Web that can access the Swiss-Prot/TrEMBL and retrieve the database. Its main sites are the ExPASy Molecular Biology website (http://www.expasy.org/) and the European Institute of Bioinformatics (EBI) website (http://www.ebi.ac.uk/swissprot/).
ExPASy is one of the main entry points of UniProtKB [5] [6] [7]. Tools in ExPASy can be used to handle several aspects of protein analysis, including BLAST search, proteomics, and sequence analysis, and to consider all splicing variants annotated in UniProtKB. A visual representation of the amino acid characteristic table developed by ExPASy enables users to see the sequence characteristics at a glance.
For more detailed information, please refer to this article [8].
5. What does the SWISS-PROT Entry Contain?
Each entry in SWISS-PROT contains the following information: Known protein sequences, references, taxonomic information, annotations, etc.
The protein entry contains a total of 14 topics.
You can use this database to query some information you need:
- Query for aliases of target proteins
- Query the subcellular localization of protein
- Query for post-translational modifications of proteins
- Query the amino acid sequence of the protein
- Query the expected molecular weight of the protein
- Query for different possible isoforms
The specific content is as follows:
5.1 Function
The content of this part is biological knowledge about protein function. In this section, you will get a detailed understanding of the biological functions of proteins and the biological processes in which they are involved. This will help you determine your research direction.
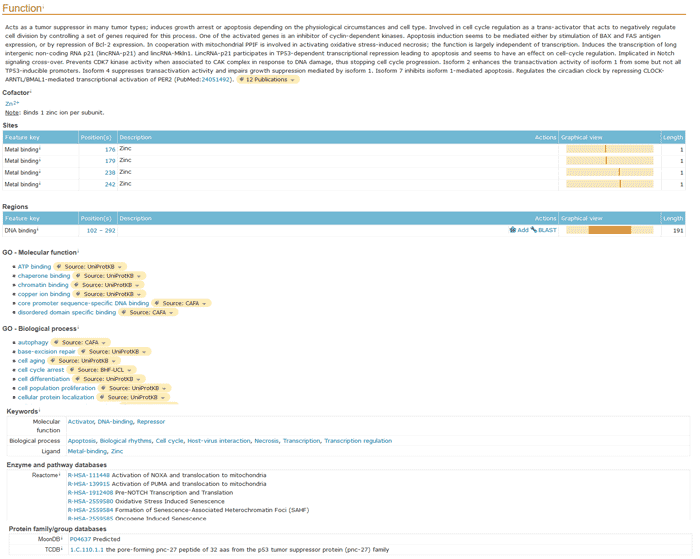
Figure 1 Function section of a UniProtKB entry.
The subsections of this section are:
- General annotations on function, catalytic activity, cofactors, enzyme regulation, biophysical and chemical characteristics and pathways.
- Graphical view of the sequence characteristics of active sites, metal binding, binding sites, sites, calcium binding, zinc fingerprinting and DNA binding.
- GO term for the molecule function.
- Key words of molecular function, biological process and ligands.
- Cross-references to the family, enzyme and pathway databases.
5.2 Names and taxonomy
This section describes the protein name, gene name and classification of the organism.
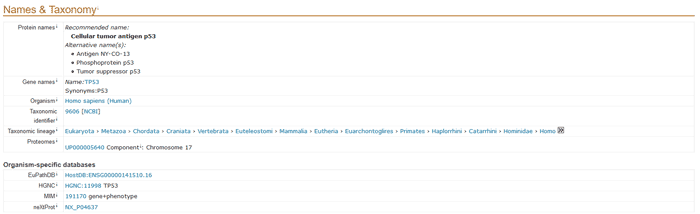
Figure 2 Names & Taxonomy section of a UniProtKB entry.
5.3 Subcellular location
Here is some information related to the biological knowledge of protein localization and topology. It mainly includes the following contents:
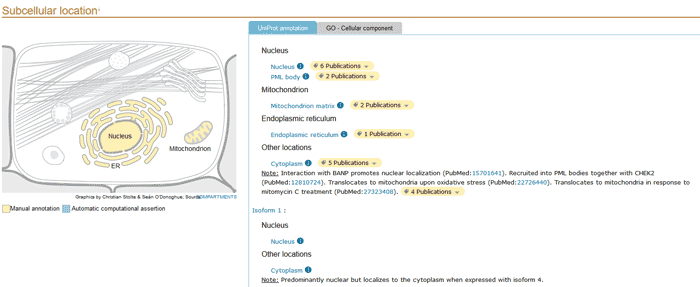
Figure 3 Subcellular location section of a UniProtKB entry.
- General annotations dealing with subcellular localization.
- Describe the sequence characteristics of transmembrane and topological domains in a graphical view.
In the “Topology”, it is possible to distinguish which sequences are extracellular, which are transmembrane, and which are intracellular! This can be used as a reference when studying its different functional segments!

Figure 4 Topology section of a UniProtKB entry.
- GO term of the cellular component. Yellow indicates manual annotation, which is more accurate; blue represents automatic computational assertion.
- Keywords for the cellular component part.
5.4 Pathology and biotech
Knowledge of the biology of disease and the phenotypes associated with protein deficiency.
This section contains the following subsection:
- General annotations on the disease, natural variation, allergic properties, biotechnological use, toxic dose and drug use associated with the entry.
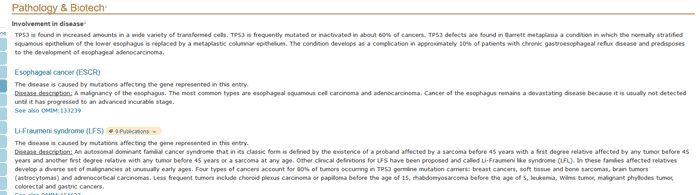
Figure 5 Pathology & Biotech section of a UniProtKB entry.
You can learn about some diseases related to this protein, which should provide a big direction for your research.
- Describe the sequence characteristics of disrupted phenotypes and mutagenesis in a graphical view.
If you know the mutation site of your protein sequence, you can query here for the possible functional changes caused by the mutation of this site, as shown in the “description” section.
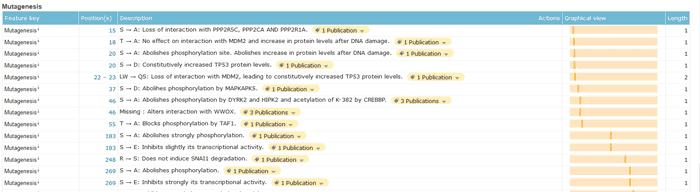
Figure 6 Pathology & Biotech section of a UniProtKB entry.
- Key words of the disease.
- Cross-reference to an organism-specific database.
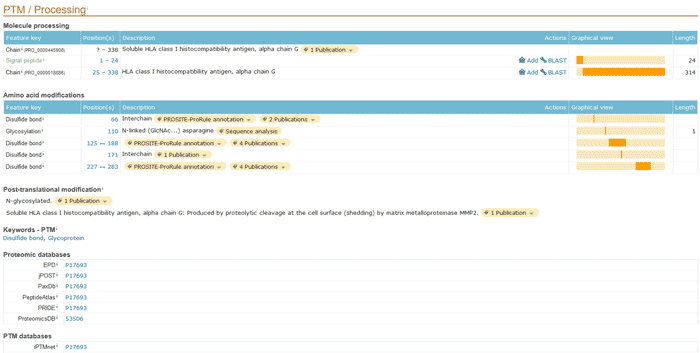
5.5 PTMs / Processing
It includes biological knowledge of post-translational modification of proteins.
The different subsections of the PTM / Processing are:
5.6 Expression
It contains information related to the biological knowledge of protein expression.
The different subsections of the expression section are:
- General annotations on tissue specificity, developmental stages and induction.
- Key words in the development stage.
- Cross-reference to the gene expression and organism-specific databases.

Figure 8 Expression section of a UniProtKB entry.
5.7 Interaction
Here is information relevant to the biological knowledge of protein interactions.
The different subsections of the interaction section are:
- General annotations of subunit structures.
- Specific annotations for binary interactions.
- Cross-reference to protein-protein interactions databases and chemistry databases.
5.8 Structure
Information related to the biological knowledge of protein structure.
It includes the following:
- Sequence characteristics of turn, beta strand and helix with a graphical view (if available).
- Cross-references to 3D structured databases and miscellaneous databases.
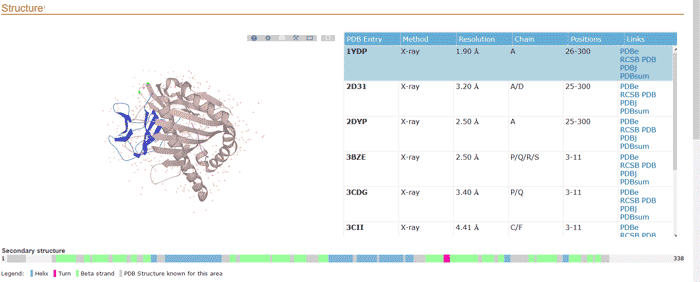
Figure 9 Structure section of a UniProtKB entry.
5.9 Family & Domains
The different subsections of the Family & Domains section are:
- Sequence features describing domain, repeat, compositional bias, region, coiled coil, motif, and domain with a graphical view.
- General annotations dealing with sequence similarity. You can know which family the protein belong to.
- Keywords for the “domain”.
- Cross-references to the phylogenomic, family and domain databases.

Figure 10 Family & Domains section of a UniProtKB entry.
5.10 Sequence
General metadata for a given sequence, such as sequence length, molecular weight, and CRC64 checksum (64-bit cyclic redundancy check value).
The different subsections of the “sequence” are:
- Sequence status: complete or fragment
- The canonical protein sequence
- Computationally mapped potential isoform sequences.
- Sequence features describing natural variant, alternative sequence, sequence uncertainty, sequence conflict, nonadjacent residues, non-terminal residue, and non-standard residue with a graphical view.
- Keywords of the ‘Coding sequence diversity’ section.
- Cross-references that point to sequence, genome annotation databases and polymorphism databases.
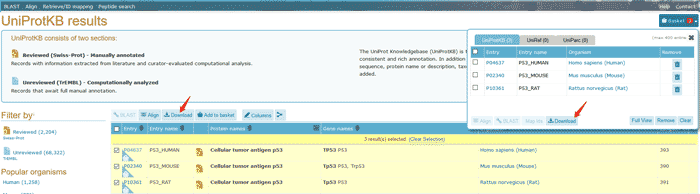
You can download the data by click FASTA button. You can also use Align tool to align this entry with its isoforms. Interested proteins can be stored in the basket by clicking the Add to basket button for later comparison or download.
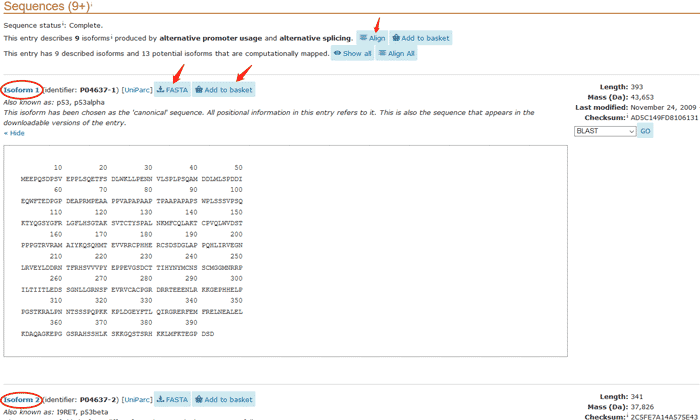
Figure 11 The canonical protein sequence and isoforms
In WB experiments, if the protein size is inconsistent with the predicted size, you can use SWISS-PROT to solve these problems:
First you need to determine whether the protein is endogenous or exogenous and whether it's a full-length sequence.
Query the SWISS-PROT database for other isoforms for this protein.
In the literature of SWISS-PROT, it was found whether there was shear activation of the protein.
Determine whether a protein is a dimer or a polymer.
Search in SWISS-PROT whether there is modification after protein expression, if there is modification will lead to protein increase. This information is available in the “PTMs/Processing” section.
5.11 Similar Proteins
This section provides links to UniRef100, UniRef90 and UniRef50, respectively, corresponding to protein sequences sharing 100%, 90% or 50% identity.
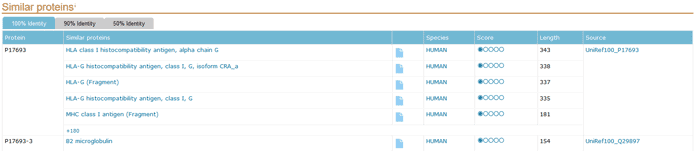
Figure 12 Similar proteins sections of a UniProtKB entry
5.12 Cross-reference
Cross-referenced sections are organized into subsections by topic. This section links the protein to several other databases that contain information about the protein. Many of these cross links are automatically added to the UniProtKB/ TrEMBL entry, but some are created manually in the UniProtKB/ SWISS-PROT entry.
The cross-reference section contains a variety of different databases:
Sequence databases; 3D structure databases; protein-protein interaction databases; chemistry databases; PTM databases; polymorphism and mutation databases; proteomic databases; protocols and materials databases; genome annotation databases; organism-specific databases; phylogenomic databases; enzyme and pathway databases; miscellaneous databases; gene expression databases; family and domain databases.
5.13 Entry Information
In addition to the primary entry number, the protein entry may contain one or more secondary entry numbers, which follow the main entry number.
5.14 Miscellaneous
- Keywords of “Technical term”.
- Documents.
In SWISSPROT, there are some tools you must know: “Blast”, “Align”, “Retrive/ID mapping”, “Peptide search”.
Blast: With the Basic Local Alignment Search Tool (BLAST), you can find regions of local similarity between sequences, which can be used to infer functional and evolutionary relationships between sequences as well as help identify members of gene families.
Align: Align two or more protein sequences with the Clustal Omega program (see also this FAQ) to view their characteristics alongside each other.
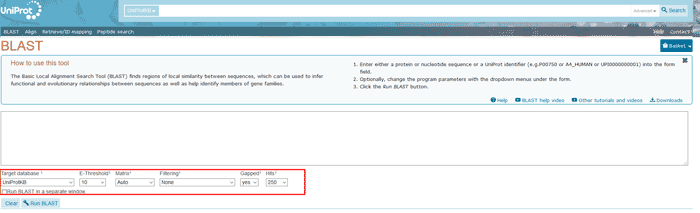
Figure 13 The BLAST tool of UniProt
Set parameters to suit your needs, default settings being: UniProtKB for the data set, 10 for the E-threshold, Matrix auto, no low complexity filtering and gap allowed.
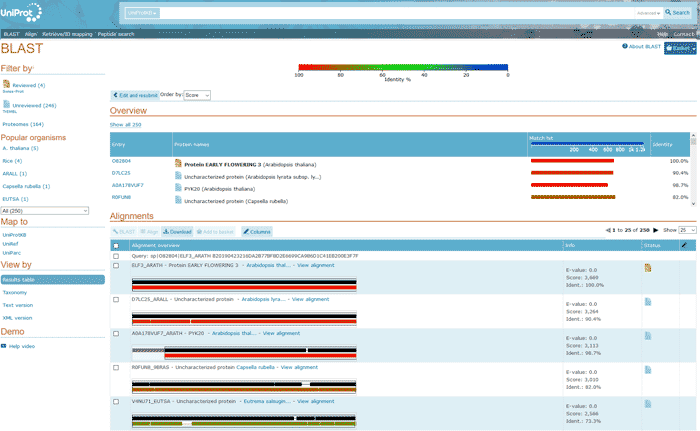
Figure 14 BLAST result
In the BLAST, the detailed list shows the matching proteins. Use different colors to show the identity between the proteins. All proteins are ranked from highest to lowest in identity.
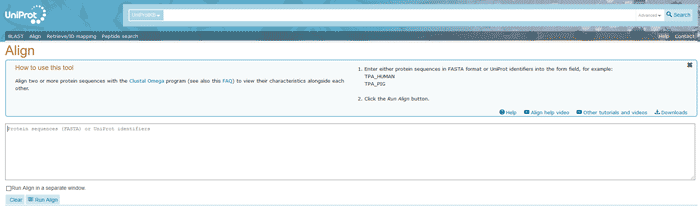
Figure 15 The Align tool of UniProt
The alignment output is interactive and gives the possibility to highlight in different colors sequence features annotated in UniProtKB as well as amino acid properties by selecting properties of interest. When more than two protein sequences are aligned, an alignment tree is also available. Alignment tree gives you an understanding of genetic relationship between these proteins.
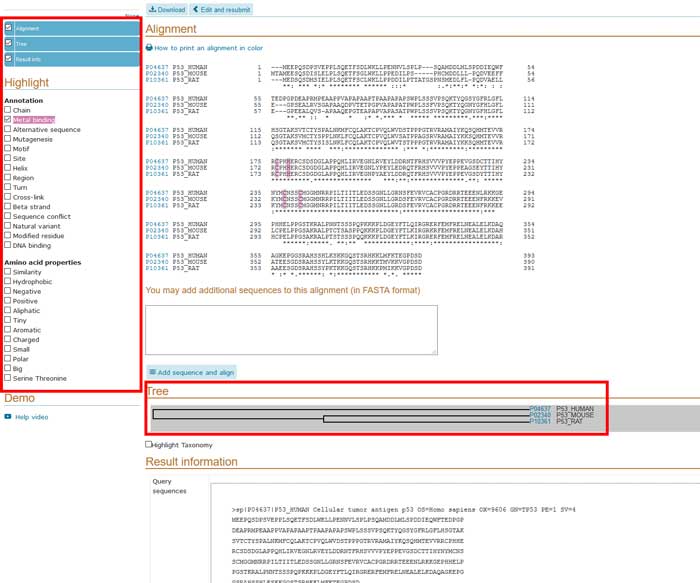
Figure 16 Protein alignment result. Partial view of the protein alignment result made on UniProtKB between P04637, P02340, P10361protein entry.
6. Display and Output of Search Results
In the detailed interface of results display, the following contents are included: general information (record name, registration number, etc.), name and source (protein name, etc.), PubMed literature information, comments, cross search, keywords, characteristics, sequence information, etc.
References
[1] Schneider M, Tognolli M, Bairoch A. The Swiss-Prot protein knowledgebase and ExPASy: providing the plant community with high quality proteomic data and tools [J]. Plant Physiology and Biochemistry (Paris), 2004, 42(12): 1013-1021.
[2] Schneider M, Bairoch A, Apweiler W R. Plant Protein Annotation in the Uniprot Knowledgebase [J]. Plant Physiology, 2005, 138(1): 59-66.
[3] Schneider M, Lane L, Boutet E, et al. The UniProtKB/Swiss-Prot knowledgebase and its Plant Proteome Annotation Program [J]. Journal of Proteomics, 2009, 72(3): 567-573.
[4] Boeckmann B. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003 [J]. Nucleic Acids Research, 2003, 31(1): 365-370.
[5] Gasteiger E. ExPASy: the proteomics server for in-depth protein knowledge and analysis [J]. Nucleic Acids Research, 2003, 31(13): 3784-3788.
[6] Walker J M. In The Proteomics Protocols Handbook [M]. Humana Press, 2005.
[7] Gattiker A, Gasteiger E, Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool [J]. Applied Bioinformatics, 2002, 1(2): 107.
[8] Boutet E, Lieberherr D, Tognolli M, et al. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View [J]. Methods in Molecular Biology, 2016, 1374: 23-54.
CUSABIO team. SWISS-PROT: When and How to Use It. https://www.cusabio.com/c-20905.html



















Comments
Leave a Comment