The COVID19 epidemic that broke out at the end of 2019 is still raging around the world. The mortality rate of SARS-CoV-2 is approximately 2.22% (as of March 5, 2021). Due to the severe threat this poses, development of antiviral drugs targeting the virus is a top priority. The team from Republic of Korea has done a study to compare the antiviral activity among Agrimonia pilosa, Galla rhois, and APRG64. Click here to view the full text of this literature…
Agrimonia pilosa (AP; Rosaceae), commonly known as hairy agrimony, is distributed widely and cultivated in Korea and China, and its aerial parts have been used as an antiviral and for treating hiatochezia, traumatic injuryand leukorrhea in Oriental medicine. Galla rhois (RG) is the gall caused by the Chinese aphid Schlechtendalia chinensis (Bell) on Rhus chinensis leaves (Anacardiaceae) and has been used in Oriental medicine for treatment of excessive sweating, persistent cough, and diarrhea.
APRG64 is a 50% EtOH aqueous extract mixture of AP and RG in a 6:4 ratio, which was optimal to exert maximum biological activities without significant pharmacological toxicity on cardiovascular, central nervous, and respiratory systems in their previous work. Therefore, they hypothesized that AP, RG, and APRG64 will display antiviral activity against SARS-CoV-2. In order to demonstrate this hypothesis, they have done this study. The results of this study have cleared the confusions as follows:
1. Does AP, RG, and APRG64 Affect the Replication of SARS-CoV-2?
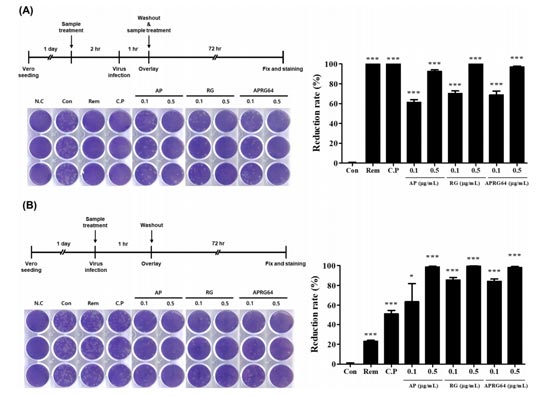
Because AP, RG, and APRG64 have clinical potential for diverse diseases, these extracts were analyzed for antiviral activity against SARS-CoV-2 firstly. Vero cells pre-treated with AP, RG, APRG64, ridesivir, or chloroquine phosphate for 2 h were infected with SARSCoV-2. At 72 h post-infection (hpi), reduction of plaque formations was assessed. Notably, all three extracts bly inhibited the formation of plaques (Fig 1A). then, Yeong-Geun Lee et al. treated vero cells infected with SARS-CoV-2 in the presence or absence of AP, RG, APRG64, ridesivir, or chloroquine phosphate. After 1 h, cells were washed three times with PBS to riove unattached viruses and extracts from cell culture media. Plaque reduction assay revealed that AP, RG, and APRG64 potently inhibited the replication of SARS-CoV-2. But ridesivir and chloroquine did not show significant antiviral activity compared with AP, RG, and APRG64 (Fig 1B). These results bly support that AP, RG, and APRG64 have potent antiviral activity against SARS-CoV-2 by interfering with viral entry through an antiviral mechanism different from that of ridesivir and chloroquine phosphate.
Fig. 1. Anti-SARS-CoV-2 activity of Agrimonia pilosa (AP), Galla rhois (RG), and their mixture (APRG64)
As mentioned before, APRG64 is a mixture of AP and RG, and because its safety in vitro as well as in vivo was proven in their previous study, its active components were investigated in the following experiment.
2. What Active Components of APRG64 Against SARS-CoV-2?
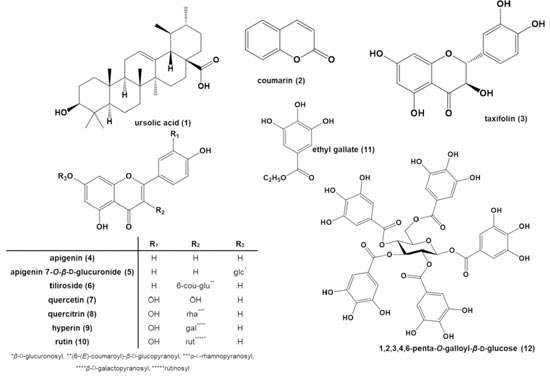
In order to discover the active components of APRG64 against SARS-CoV-2, they repeated c.c. of these plants and Isolated many components, including ursolic acid (1), ethyl gallate (11), and 1,2,3,4,6-penta-O-galloyl-β-D-glucose (12). Based on LC/MS profiling results of APRG64, one triterpenoid (1), one coumarin (2), eight flavonoids (3–10), and two gallate derivatives (11 and 12) were selected as active components of APRG64 and potential anti-COVID-19 agents (Fig. 2).
Fig. 2. Chemical structures of isolated constituents from mixture of Agrimonia pilosa (AP) leaves and Galla rhois (RG) fruits in 50% EtOH extract (APRG64)
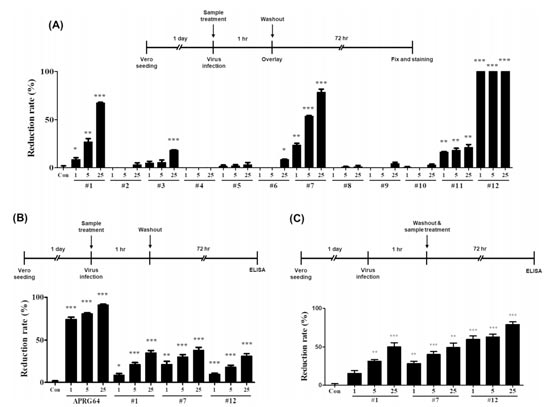
Moreover, they also investigated whether secondary metabolites isolated from APRG64 inhibit the replication of SARS-CoV-2. Vero cells were infected with SARS-CoV-2 and treated with compounds 1–12, remdesivir, or chloroquine phosphate for 1 h. Cells were washed to eliminate unattached viral particles and compounds. At 72 hpi, the number of plaques was estimated using a plaque assay. As shown in Fig. 3A, ursolic (1), quercetin (7), ethyl gallate (11), and 1,2,3,4,6-penta-O-galloyl-β-Dglucose (12) significantly inhibited the formation of plaques at all three concentrations (1, 5, and 25 µg/mL). However, compared with others, ethyl gallate (11) slightly inhibited the formation of plaques (reduction rate: 21.05% at 25µg/mL). Similar results were obtained when cell supernatants were analyzed for SARS-CoV-2 spike protein (Fig. 3B). Ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloylβ-D-glucose (12) significantly reduced the amount of SARS-CoV-2 spike proteins in cell supernatants.
Finally, they explored whether post-treatment with ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-Dglucose (12) was capable of inhibiting SARS-CoV-2 replication. As the Fig. 3C shows, post-treatment with samples significantly reduced SARS-CoV-2 spike proteins in the supernatants.
Fig. 3. Antiviral activity of active components isolated from APRG64 against SARS-CoV-2. Vero cells were seeded 1 day before infection
Collectively, the results strongly support that active components of APRG64, ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-Ogalloyl-β-D-glucose (12), inhibit SARS-CoV-2 replication by interfering with viral absorption and post-absorption stages.
3. Can the Components of APRG64 Hinder the Binding of Viral Spike RBD to Host Cells?
As you know, the binding of the viral spike RBD and ACE2 proteins on the target cell is a critical step for SARS-CoV-2 to enter the cell. Because APRG64 and its components were shown to interfere with viral absorption, the binding of the viral spike RBD onto host cells could be impeded by APRG64 components.
To test this hypothesis, molecular docking analysis of the viral spike RBD onto the structures of three major antiviral components of APRG64, ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-D-glucose (12), was performed. However, the spike RBD of SARS-CoV-2 continuously undergoes mutations, thus, the binding affinity of APRG64 components to the SARS-CoV-2 spike RBD as well as its variant B.1.1.7 spike RBD was analyzed.
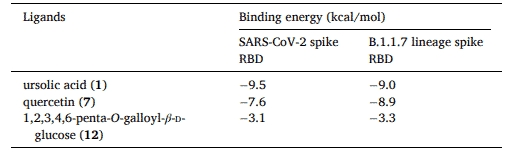
First, the B.1.1.7 lineage spike RBD structure model was generated and validated by Global Model Quality Estimation (GMQE) and Qualitative Model Energy ANalysis (QMEAN). Next, comparative molecular docking analyses of the SARS-CoV-2 spike RBD and B.1.1.7 lineage spike RBD with ursolic acid (1), quercetin (7), and 1,2,3,4,6-penta-O-galloyl-β-D-glucose (12) were performed to assess the potential antiviral effects of the active APRG64 components (Table 1).
Table 1. Binding energy calculated from molecular docking analysis of SARS-CoV-2 spike receptor-binding domain (RBD) and B.1.1.7 lineage spike RBD with three antiSARS-CoV-2 compounds of APRG64, ursolic acid (1), quercetin (7), and 1, 2, 3, 4, 6-penta-O-galloyl-β-D-glucose (12)
The data in table 1 shows, ursolic acid (1) showed the highest binding energy (− 9.5 kcal/mol and − 9.0 kcal/mol, respectively) against the SARS-CoV-2 spike RBD and B.1.1.7 lineage spike RBD compared with the other compounds. Moreover, they also forecasted the molecular binding of SARS-CoV-2 spike receptor-binding domain (RBD) and B.1.1.7 lineage spike RBD with three active components of APRG64, ursolic acid (1), quercetin (7), and 1, 2, 3, 4, 6-penta-O-galloyl-β-D-glucose (12) by Autodock. These data indicate that APRG64 has significant potential to target both SARS-CoV-2 and B.1.1.7 lineage and exert antiviral effects.
In a word, this study indicates APRG64 as a potent drug candidate to treat SARS-CoV-2 and its variants. And APRG64 is going to be a novel candidate antiviral agent to treat COVID-19, and further clinical trials are warranted.
4. Reagents Used in this Research
The key part of this research used the SARS-CoV-2 spike RBD Nanobody (CSB-RA33245A2GMY) provided by CUSABIO to detect and analyze the SARS-CoV-2 spike proteins in cell supernatants from virus-infected, sample-treated cells.
The Validated data of SARS-CoV-2 spike RBD Nanobody
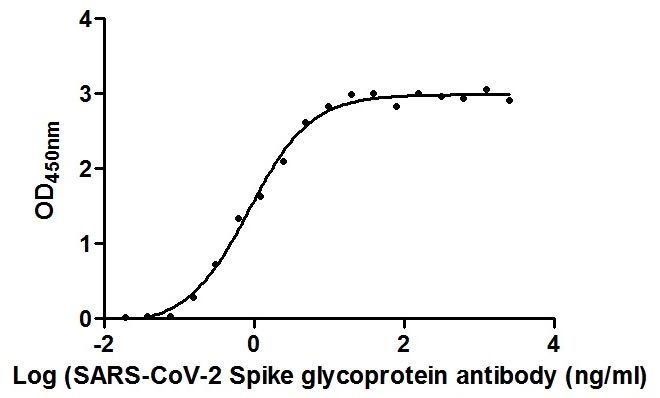
>>The Binding Activity of SARS-CoV-2 Spike RBD Nanobody with SARS-CoV-2-S1-RBD
Functional ELISA
Immobilized SARS-CoV-2-S1-RBD (CSB-YP3324GMY1) at 2 μg/ml can bind SARS-CoV-2 Spike RBD Nanobody, the EC50 is 0.8674 ng/ml.
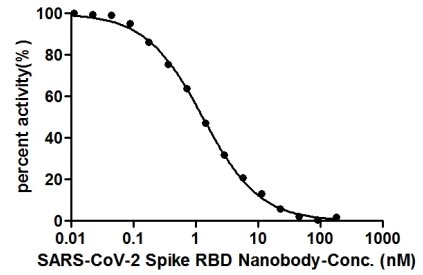
>>The Binding Activity of SARS-CoV-2 Spike RBD Nanobody was detected by competing with ACE2-HRP conjugate for binding to SARS-CoV-2-S1-RBD
The binding signal of SARS-CoV-2-S1-RBD (CSB-YP3324GMY1) and ACE2-HRP (CSB-MP866317HU) conjugate was gradually reduced as the SARS-CoV-2 Spike RBD Nanobody concentrations increased. It indicated that this SARS-CoV-2 Spike RBD Nanobody effectively inhibited the SARS-CoV-2-S1-RBD/ACE2 binding. And the IC50 of this SARS-CoV-2 Spike RBD Nanobody is 1.296 nM.
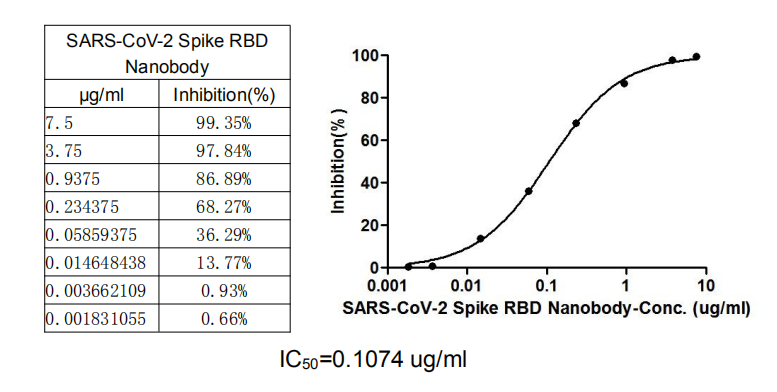
>>The neutralizing role of SARS-CoV-2 Spike RBD Nanobody is detected by competitively preventing SARS-CoV-2-S1-RBD from binding to ACE2-HRP conjugate
The inhibition efficacy of the SARS-CoV-2-S1-RBD /ACE2 binding was positively proportionally to the SARS-CoV-2 Spike RBD Nanobody concentrations. It showed that this SARS-CoV-2 Spike RBD Nanobody effectively inhibited the SARS-CoV-2-S1-RBD/ACE2 binding. And the IC50 of this SARS-CoV-2 Spike RBD Nanobody is 0.1074 μg/ml.
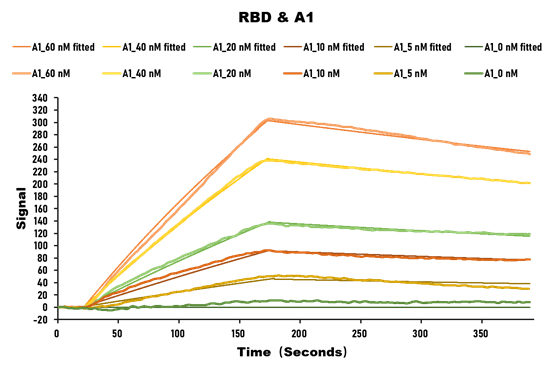
>>The activity of SARS-CoV-2 Spike RBD Nanobody is detected by LSPR
SARS-CoV-2 Spike protein RBD His/Sumostar Tag (CSB-YP3324GMY1) captured on COOH chip binding to the SARS-CoV-2 Spike RBD Nanobody, increases the local refractive index (RI), leading to a red shift of the LSPR peak position. The detected affinity constant of SARS-CoV-2 Spike protein RBD/SARS-CoV-2 Spike RBD Nanobody binding is 28.2nM.
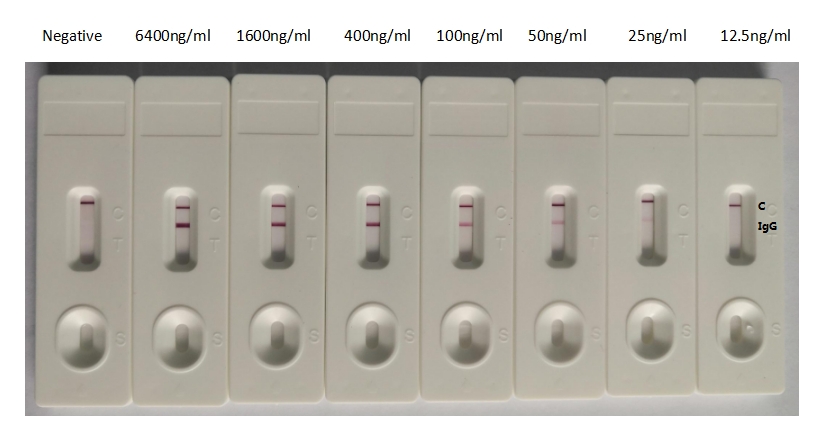
>>Colloidal Gold Immunochromatography Assay Validation
GICA
In the GICA detection system, the background of antibody (CSB-RA33245A2GMY) is clean, the detection limit can be as low as 25ng/ml (1.75ng/0.07ml), and the sensitivity is very good.
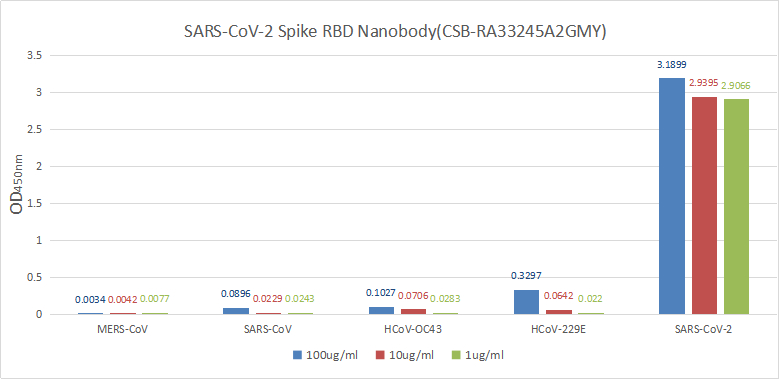
>>Enzyme Linked Immunosorbent Assay Validation
Immobilize various SARS proteins at concentration of 2μg/ml on solid substrate, then react with the nanobody at concentration of 100μg/ml, 10μg/ml and 1μg/ml. It shows this nanobody is specific for SARS-CoV-2-S1-RBD protein, without any cross-reactivity with MERS-CoV, SARS-CoV, HCoV-OC43 or HCoV-229E.
CUSABIO team. Agrimonia pilosa, Galla rhois, and APRG64, Which has the best antiviral activity against SARS-CoV-2?. https://www.cusabio.com/c-21041.html




Comments
Leave a Comment