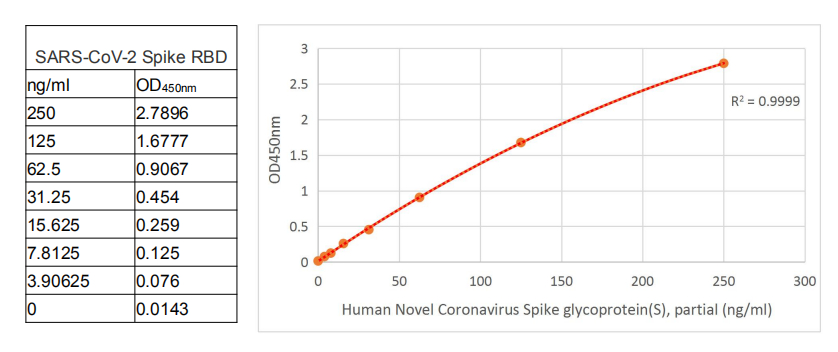
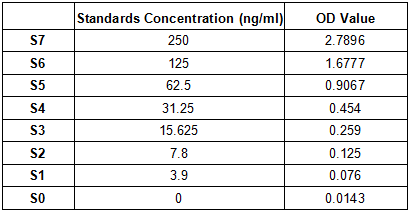
Cusabio supplies the following matched antibody pair for detecting SARS-CoV-2 Spike protein in Sandwich ELISA assay. Cusabio guarantees that the antibody works for the applications/species stated on our website and datasheet, a replacement or refund will be provided if the antibody does not work as described on our datasheet.
CSB-EAP33245 SARS-CoV-2 Spike RBD Antibody Pair 1 https://www.cusabio.com/Antibody-Pairs/S-Antibody-Pair-12928623.html
Concentration:
Capture Antibody: 1mg/ml
Detection Antibody: 0.42mg/ml
If you also need a standard protein, we would like to recommend the following product.
CSB-MP3324GMY1b1 https://www.cusabio.com/Recombinant-Protein/Recombinant-Human-Novel-Coronavirus-Spike-glycoprotein-S---partial--12928577.html