The COVID-19 outbreak has caused infection in a large number of people since it was discovered in late 2019. With the increase in the number of infected people and the continuation of the epidemic, SARS-CoV-2 continues to evolve and mutate, producing a large number of variants. Some of these SARS-CoV-2 variants have become significantly more transmissible and pathogenic, and some have shown antigenic escape.
These variants are of concern to public health departments and the public. In many news reports, we have seen various names for SARS-CoV-2 variants, such as "South African variant", B.1.617.2, Delta, Omicron and various other names.
So how did SARS-CoV-2 variants get their names?
In general, there are four nomenclature methods for the currently available SARS-CoV-2 variants: Pango, GISAID, Nextstrain, and Greek letters.
The following are the names of a few of the more highly regarded SARS-CoV-2 variants for each nomenclature.
|
WHO label
|
Pango lineage
|
GISAID clade
|
Nextstrain clade
|
Earliest documented samples
|
|
Alpha
|
B.1.1.7
|
GRY
|
20I (V1)
|
United Kingdom, Sep-2020
|
|
Beta
|
B.1.351
|
GH/501Y.V2
|
20H (V2)
|
South Africa, May-2020
|
|
Gamma
|
P.1
|
GR/501Y.V3
|
20J (V3)
|
Brazil, Nov-2020
|
|
Delta
|
B.1.617.2
|
G/478K.V1
|
21A, 21I, 21J
|
India, Oct-2020
|
|
Omicron
|
B.1.1.529
|
GR/484A
|
21K, 21L, 21M, 22A, 22B, 22C, 22D
|
Multiple countries, Nov-2021
|
Table1: Names of SARS-CoV-2 variants (Source: WHO)
Note:
According to the level of attention, mutated viruses are divided into 4 types [1]:
Variants being monitored (VBM)
Variants of interest (VOI)
Variants of Concern (VOC)
Variants of high consequence (VOHC)
This classification is not fixed. Variants, like Alpha, Beta, Gamma, and Delta in the above table, have been designated as VOCs. But they are currently designated as VBM. Omicron is still a VOC.
Currently, no SARS-CoV-2 variants are designated as VOI.
Now, let's talk about the characteristics of each naming system in detail.
1. WHO Label (the Greek letters)
Figure 1: Greek Alphabet Letters (Source: Immediate Media)
In May 2020, a new variant of SARS-CoV-2, B.1.351, was discovered in South Africa. Some scientists call it GH/501Y.V2, and some called it 20H/S:501Y.V2. There were media, which directly called the virus "South African variant". For a time, the names of the SARS-CoV-2 variants were varied. And the same thing happened to India in October 2020.
Given the complexity of naming SARS-CoV-2 variants, as well as the stigma and discrimination caused by regional naming, WHO released the Greek letter name of SARS-CoV-2 variants on May 31, 2021.
Not all mutants are named with Greek letters. Only those high-profile virus variants have Greek-letter names.
In view of the complexity of naming SARS-CoV-2 variants, as well as the stigma and discrimination caused by naming by region, the WHO released the Greek letter names for SARS-CoV-2 variants on 31 May 2021.
It is important to note that not all variants are named with the Greek alphabet. Only those variants that are of high interest will have Greek letter names.
Of course, not all Greek letters are used in order for the naming of the virus variants. There are 24 Greek letters in total, 13 of which have been used so far. From Alpha to Omicron, two of these letters are not used in the naming of SARS-CoV-2 variants.
Why have these letters not been named?
It has been explained that "nu" is pronounced like "new" and therefore has not been used; "Xi" is pronounced the same as a Chinese surname, for example, the surname of Chinese President Xi Jinping is Xi. Therefore, Xi has not been used for the naming of new crown variants. These explanations make sense to a great extent.
The Greek alphabet naming is a lot more convenient for the public. For example, the variant, Omicron, was named B.1.1.529 before it received its Greek letter name. When listening to the radio, you hear B-1-1-529, which can be confusing.
However, there is still a problem with the Greek letter names.
What to do when you run out of Greek letters?
The WHO official, Van Kerkhove, said WHO was considering naming variants after constellations. However, this naming rule has not yet been officially activated, although the constellation nickname of the variants is already being used on social media. For example, the subtype BA.2.75 of Omicron is known on social media as Centaurus and has been widely disseminated.
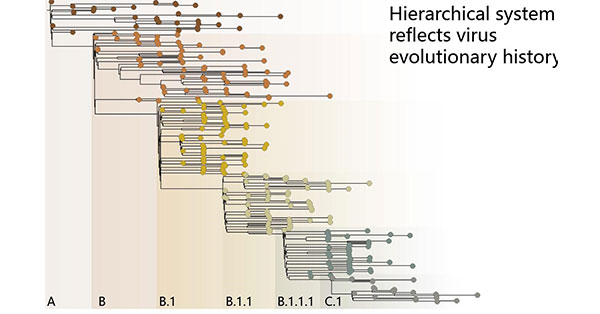
2. Pango lineage
Figure 2: Pango lineages (Source: pango.network)
The Pango nomenclature is based on a phylogenetic hierarchy that clearly indicates the evolutionary position of the variant and how distantly related it is to variants. Under this rule, the name consists of a capital letter prefix and a numerical suffix separated by a dot. Any letter of the Latin alphabet may be used (alone or in combination) within the prefix (except I, O and X).
Delta is called B.1.617.2 under the Pango naming convention, and Omicron is called B.1.1.529.
Let’s check the structure of the name.
The letter, "B", represents the lineage to which the variant belongs among the many variants, the following number represents the serial number of the lineage and the descendant lineage, with each dot indicating hierarchical relationships.
For example lineage B.1.1.7 is the seventh descendant of lineage B.1.1, while B.1.1 is the first descendant of lineage B.1.
In addition, to avoid long lineage labels, there are up to three levels of numeric suffixes (primary, secondary, and tertiary). Descendants of the third suffix use the next letter prefix in the alphabet to act as an alias for the parent lineage name.
For example, the first descendant of lineage B.1.1.1 is not named B.1.1.1.1, but C.1, so the prefix C acts as an alias for B.1.1.1.
According to the rules, details of each alias should be provided in the current list of genealogical descriptions so that the ancestral relationships of all Pango lineage can be clearly reconstructed.
For example, the reference to BF.1 is usually followed by the label "Alias of B.1.1.529.5.2.1.1". [2]
Tips:
How to read the Pango names?
The dot in the middle of the name is very important in the pronunciation of the Pango name, but it is often overlooked. For example, B.1.17 and B.1.1.7.
B.1.17 is read as B, 1, 17.
B.1.1.7 is read as B, 1, 1, 7.
The dot acts as a pause.
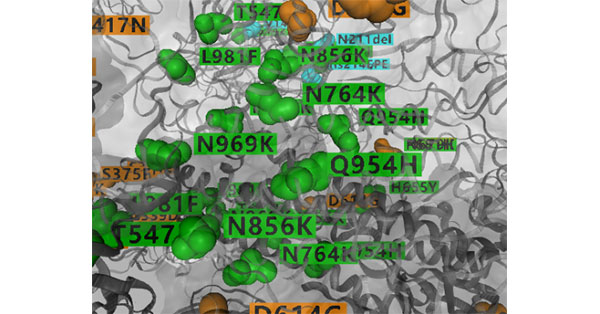
3. GISAID
Figure 3: Partial 3D Structure of Spike with Amino Acid Changes (Source: GISAID)
GISAID, first released in May 2008, is a data-sharing platform for epidemic viruses and has been recognized by the WHO and the European Union.
The SARS-CoV-2 variants information displayed by GISAID is mainly named by the amino acid variation corresponding to the virus mutation site. For example, one of the previously rampant Delta viruses strains around the world, G/478K.V1.
GISAID is one of the nomenclature systems for SARS-CoV-2 variants, which uses the amino acid variants corresponding to the mutation site of the virus to name the SARS-CoV-2 variant. For example, one of the previously rampant variant of Delta viruses worldwide, G/478K.V1.
Let's take a look at G/478K.V1.
The "G" in the name is the variant branch name. The branch names of nine SARS-CoV-2 variants are given in GISAID: S, L, V, G, GK, GH, GR, GV, and GRY . [3]
"478K" is the mutation site, where the amino acid at position 478 is mutated to lysine.
"V1" represents the first of the Delta variant to be identified. [4]
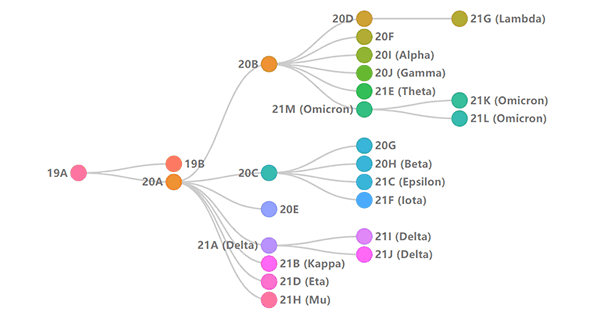
4. Nextstrain
Figure 4: Illustration of phylogenetic relationships of SARS-CoV-2 clades (Source: Nextstrain)
Nextstrain is an open-source project that exploits pathogen genome data. In the Nextstrain system, the naming method of the SARS-CoV-2 variants is slightly different from that of GISAID. It will start with the year, and then name the virus mutation branches that appear in the year in the order of the English alphabet.
Take Omicron as an example, its Nextstrain branch contains 21K, 21L, 21M, 22A, 22B, 22C, 22D. Among them, there are about 19 mutation points of 21K, which include S:R346K, S:A701V, S:N211R, S:I1081V, S:A67-, S:I68-, etc. [5]
So you may have seen variant names like this, 21K/ S:R346K, which tells you the amino acid, arginine (R), at position 346 on the S protein is mutated to lysine (K) in the clade of 21K.
Conclusion
The four SARS-CoV-2 variants have their own significance. Either it is easy for the public to understand, or it is conducive to the classification of scientific research. They will all play a major role in humanity's fight against COVID-19.
References
[1] Tracking SARS-CoV-2 variants. World Health Organization.
https://www.who.int/activities/tracking-SARS-CoV-2-variants
[2] SARS-CoV-2 lineages
https://cov-lineages.org/lineage_list.html
[3] GISAID Statements & Clarifications
https://gisaid.org/resources/statements-clarifications/clade-and-lineage-nomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/
[4] Garcia-Beltran WF, Lam EC, et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. medRxiv. Update in: Cell. 2021, Apr 29.
[5] Variant: 21K (Omicron)
https://covariants.org/variants/21K.Omicron
CUSABIO team. How did SARS-CoV-2 Variants get Names?. https://www.cusabio.com/c-21080.html





Comments
Leave a Comment