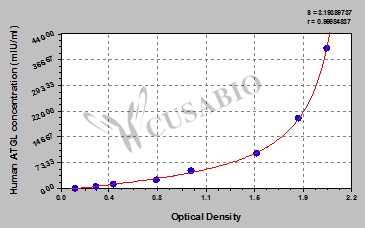
This human ATGL ELISA kit employs the quantitative sandwich enzyme immunoassay technique to measure the levels of human ATGL in different samples, including serum, urine, or tissue homogenates. The enzyme-substrate chromogenic reaction is also used to amplify the signal and quantify the levels of the analyte through the intensity of the colored product. The color intensity positively correlates with the amount of ATGL bound in the initial step.
ATGL is a triglyceride lipase that catalyzes the initial step in adipose triglyceride (TG) lipolysis, cooperating with other enzymes to hydrolyze TG to produce fatty acids (FAs) that are crucial energy substrates, precursors for the generation of membrane lipids, and ligands of nuclear receptors. It plays an important role in whole-body energy homeostasis. In addition to mobilizing TG for energy production, ATGL also affects obesity and adipose tissue inflammation, heart failure protection, glucose homeostasis, as well as thermoregulation.