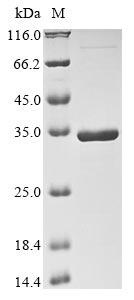
Recombinant Amanita muscaria DOPA 4,5-dioxygenase (DODA) is expressed in E. coli and contains the complete protein sequence spanning amino acids 1 to 228. The protein has been designed with an N-terminal 10xHis-tag and a C-terminal Myc-tag to help with purification and detection. SDS-PAGE analysis confirms the product achieves greater than 90% purity, which appears to make it appropriate for various research experiments.
DOPA 4,5-dioxygenase (DODA) is an enzyme that catalyzes the oxidative cleavage of DOPA—a step that may be critical in certain metabolic pathways. This activity seems particularly important in producing various secondary metabolites, though the specific roles in biochemical and pharmacological contexts remain areas of active investigation. Research into DODA could offer insights into enzymatic mechanisms and help researchers better understand how metabolic pathways function in fungi.
Potential Applications
Note: The applications listed below are based on what we know about this protein's biological functions, published research, and experience from experts in the field. However, we haven't fully tested all of these applications ourselves yet. We'd recommend running some preliminary tests first to make sure they work for your specific research goals.
Based on the provided information, the recombinant DODA protein may not be correctly folded or bioactive without experimental validation. DODA is an enzyme that requires precise folding for its dioxygenase activity, including proper coordination of iron cofactors and substrate binding. The E. coli expression system may not fully support eukaryotic-like folding or post-translational modifications needed for optimal activity. The dual tags (N-terminal 10xHis and C-terminal Myc) could sterically hinder the protein's structure, particularly if the tags interfere with active site accessibility or oligomerization. The purity >90% indicates low contaminants, but does not guarantee correct folding. Without validation (e.g., enzyme activity assays with L-DOPA, circular dichroism for secondary structure), the protein's folding and bioactivity remain uncertain. Therefore, while E. coli can express functional enzymes, the dual tags, and a lack of validation poses risks.
1. In Vitro Enzyme Activity Characterization
This recombinant DODA protein can be used for enzyme activity studies only if correct folding and catalytic function are experimentally verified. The dual tags may alter the active site conformation, leading to inaccurate kinetic parameters (Km, Vmax). If misfolded, substrate specificity assays may yield false results. It is recommended to first validate dioxygenase activity using L-DOPA as a substrate and remove tags if possible for reliable measurements. The purity >90% supports quantitative assays but does not ensure functional integrity.
2. Antibody Development and Validation
This recombinant DODA can generate antibodies, but the dual tags may dominate the immune response, resulting in antibodies that primarily recognize tags rather than DODA-specific epitopes. If misfolded, antibodies may not recognize native DODA in fungal systems. For reliable outcomes, validate antibodies using tag-free DODA or confirm specificity with Amanita muscaria extracts. The Myc tag provides a detection control but does not guarantee utility for native protein studies.
3. Protein-Protein Interaction Studies
The dual-tagged DODA can be employed in pull-down assays, but interactions may be tag-mediated or non-specific if the protein is misfolded. The tags allow immobilization and detection, but biologically relevant binding partners in betalain pathways may not bind without native conformation. Validate folding first and include controls (e.g., tag-only proteins) to minimize artifacts. Results should be confirmed with complementary methods.
4. Structural and Biophysical Analysis
This recombinant DODA is unsuitable for high-resolution structural studies (e.g., crystallography or NMR) without tag removal and folding validation. The tags introduce flexibility and heterogeneity, hindering data interpretation. Biophysical techniques (e.g., circular dichroism) can assess general folding but may be confounded by tag contributions. For meaningful insights, remove tags and confirm native-like structure through activity assays.
5. Comparative Enzyme Evolution Studies
This DODA protein can be used for comparative studies only if properly folded and active. Misfolded DODA may not accurately represent evolutionary relationships or functional differences. Validate enzyme activity and structure before comparing with homologs from other species. The standardized expression is beneficial but requires functional confirmation for reliable conclusions.
Final Recommendation & Action Plan
To ensure reliable results, first validate the folding and bioactivity of the recombinant DODA protein using enzyme activity assays with L-DOPA to confirm catalytic function, circular dichroism to assess secondary structure, and size-exclusion chromatography to check oligomeric state. Given the potential interference from dual tags, remove them via proteolytic cleavage (if cleavage sites are available) and re-purify the tag-free protein for functional and structural studies. For applications like antibody development, use the current protein but characterize antibodies against tag-free DODA or fungal extracts. In all cases, include appropriate controls, such as tag-only proteins, inactive mutants, or known DODA standards, to account for folding-related artifacts. Prioritize validation before quantitative or comparative applications to avoid misleading data.