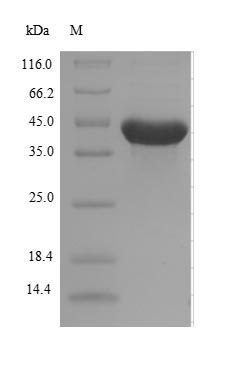
Recombinant Escherichia coli Alpha-ketoglutarate-dependent dioxygenase AlkB gets produced in E. coli expression systems and carries both an N-terminal 10xHis-SUMO tag and a C-terminal Myc tag. The protein spans its complete length, covering amino acids 1 to 216, and reaches a purity level greater than 90% as determined by SDS-PAGE. This high-quality preparation is meant for research use only.
E. coli's Alpha-ketoglutarate-dependent dioxygenase AlkB appears to play a key role in repairing alkylated nucleic acids—a process that seems critical for maintaining genetic stability. The enzyme catalyzes the oxidative demethylation of alkylated bases, which essentially reverses damage caused by environmental and endogenous alkylating agents. Studying this enzyme may offer important insights into bacterial DNA repair mechanisms and could shed light on broader molecular pathways involved in cellular maintenance and repair.
Potential Applications
Note: The applications listed below are based on what we know about this protein's biological functions, published research, and experience from experts in the field. However, we haven't fully tested all of these applications ourselves yet. We'd recommend running some preliminary tests first to make sure they work for your specific research goals.
Based on the provided information, the recombinant E. coli AlkB protein is expressed in its native E. coli system, which significantly increases the probability of proper folding and functionality. As a bacterial enzyme expressed in its homologous prokaryotic system, AlkB has a high likelihood of correct folding since E. coli contains the necessary cellular machinery for its own proteins. The protein is full-length (1-216aa) with dual tags (N-terminal 10xHis-SUMO and C-terminal Myc) and exhibits high purity (>90%). The SUMO tag may enhance solubility and proper folding. However, since activity is explicitly unverified and the presence of dual tags may potentially interfere with the active site or substrate access, the protein cannot be assumed to be fully functional without experimental validation of its dioxygenase activity.
1. Protein-Protein Interaction Studies Using Pull-Down Assays
The dual tagging system enables technical feasibility for interaction studies. The homologous expression system favors proper folding, which increases confidence in the physiological relevance of identified interactions. However, the tags may still cause steric hindrance or non-specific binding. This application should include appropriate controls (e.g., tag-only controls) to distinguish specific from artifactual interactions.
2. Antibody Development and Validation
The recombinant AlkB can serve as an effective immunogen for generating antibodies against linear epitopes. The high purity supports immunization protocols. However, antibodies may not recognize conformational epitopes if tags affect protein folding. Validation against native E. coli AlkB is recommended.
3. Biochemical Characterization and Enzyme Kinetics Studies
This application is well-suited but requires activity validation for kinetic studies. Basic biochemical characterization is feasible, but enzyme kinetics parameters (Km, Vmax) should only be determined after confirming dioxygenase activity. The tags may affect catalytic efficiency, so results should be interpreted with appropriate controls.
4. Protein Stability and Folding Studies
This application is highly appropriate. Biophysical techniques (circular dichroism, differential scanning calorimetry) can directly assess the protein's folding state and stability. These studies are valuable for characterizing the recombinant E. coli AlkB protein and can inform about its suitability for other applications.
Final Recommendation & Action Plan
Given the homologous expression system, this AlkB has high potential for proper folding and function. Recommended first steps: 1) Validate dioxygenase activity using standard assays with appropriate substrates (methylated DNA/RNA) and cofactors (α-ketoglutarate, Fe²?, ascorbate); 2) Perform biophysical characterization (size-exclusion chromatography, circular dichroism) to confirm proper folding; 3) If possible, compare with tag-cleaved protein to assess tag effects. Antibody development and stability studies can proceed immediately. For interaction studies, include appropriate controls to validate physiological relevance. For reliable kinetic parameters, use activity-verified protein and include known substrates as positive controls.