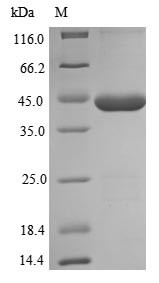
Recombinant Escherichia coli dITP/XTP pyrophosphatase (rdgB) is expressed in E. coli and covers the full length of the protein (1-197 amino acids). This product includes an N-terminal 10xHis-SUMO tag and a C-terminal Myc tag, which help with purification and detection. The protein appears to be purified to over 85% purity, as determined by SDS-PAGE, suggesting it should work well for research applications.
dITP/XTP pyrophosphatase is an essential enzyme in Escherichia coli that's involved in nucleotide metabolism. It breaks down non-canonical nucleotides like dITP and XTP to prevent their incorporation into DNA, which helps maintain genomic stability. This function makes it an important subject for studying DNA replication fidelity and how cells respond to nucleotide pool imbalances.
Potential Applications
Note: The applications listed below are based on what we know about this protein's biological functions, published research, and experience from experts in the field. However, we haven't fully tested all of these applications ourselves yet. We'd recommend running some preliminary tests first to make sure they work for your specific research goals.
E. coli rdgB is a bacterial enzyme that hydrolyzes non-canonical nucleotides (dITP/XTP) to prevent their incorporation into DNA. The E. coli expression system is compatible with this bacterial protein, increasing the likelihood of correct folding. However, the dual N-terminal His-SUMO tag (∼20 kDa) and C-terminal Myc tag may sterically interfere with the enzyme's active site or oligomerization. While the full-length protein (1-197aa) contains all functional domains, the large tags may compromise enzymatic activity. The probability of correct folding is moderate, but functional activity requires experimental validation due to potential tag interference.
1. Protein-Protein Interaction Studies Using Tag-Assisted Pull-Down Assays
Protein-protein interactions require a precise tertiary structure that may be compromised by the large dual tags. The large N-terminal SUMO tag and C-terminal Myc tag may sterically block interaction interfaces or cause misfolding, leading to non-specific binding or failure to identify genuine partners (e.g., DNA repair enzymes). If correctly folded, it could reveal physiological interactions, but results require validation with tag-free protein.
2. Antibody Development and Validation Platform
This recombinant rdgB serves as an excellent immunogen for generating antibodies against E. coli rdgB. The full-length sequence ensures comprehensive coverage of the epitope. The dual tags provide additional epitopes for screening and controls for specificity testing. These antibodies will be valuable for detecting rdgB in bacterial samples.
3. Biochemical Characterization and Substrate Specificity Analysis
This is the critical first step to assess protein quality and activity. Techniques like size-exclusion chromatography can determine oligomeric state, while enzymatic assays with dITP/XTP substrates can validate pyrophosphatase activity. However, the tags may sterically hinder substrate access to the active site, potentially yielding false-negative results in activity assays. These analyses are essential but must include activity validation to confirm functional integrity despite the tags.
4. Structural and Biophysical Studies
This application is suitable for basic biophysical characterization but is limited to structural studies. Techniques like circular dichroism can analyze secondary structure, but the large tags will dominate spectroscopic signals and prevent accurate analysis of the native rdgB structure. SEC-MALS can assess oligomeric state but may be affected by tag interference.
Final Recommendation & Action Plan
This dual-tagged recombinant rdgB is reliable for antibody development but requires rigorous activity validation before functional studies due to potential tag interference. The immediate priority is Application 3 (Biochemical Characterization) to test enzymatic activity with dITP/XTP substrates and assess oligomeric state via SEC-MALS. If activity is confirmed despite the tags, proceed cautiously with Application 1 (Interaction Studies). Application 2 (Antibody Development) can proceed immediately. Application 4 should be limited to basic biophysical properties. For reliable structural or detailed functional studies, tag removal or use of tag-free rdgB is recommended. This systematic approach ensures appropriate use based on functional validation.