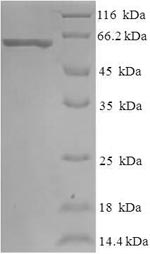
The production of this recombinant E.coli degP protein is just like all recombinant proteins. The process involved transfecting E.coli cells with DNA vector containing the template of recombinant DNA. The E.coli cells containing the template were then cultured so that they could transcribe and translate the degP protein. N-terminal 6xHis-SUMO tag was used in the process. The purity is 90% determined by SDS-PAGE.
degP is a gene providing instructions for making a protein called periplasmic serine endoprotease DegP (also known as heat shock protein DegP or protease Do). DegP protein belongs to the peptidase S1C family. DegP is a heat shock protein induced in response to pac overexpression, suggesting that the protein could possibly suppress the physiological toxicity caused by pac overexpression. Increasing evidence indicates that the production of soluble recombinant penicillin acylase in Escherichia coli can be enhanced via coexpression of a periplasmic protease/chaperone, DegP.