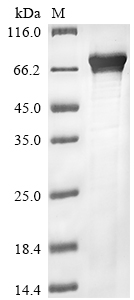
Amino acids 1-593 constitute the expression domain of recombinant Human CRY2. The calculated molecular weight for this CRY2 protein is 74.4 kDa. This CRY2 protein is produced using e.coli expression system. Fusion of the N-terminal 10xHis tag and C-terminal Myc tag into the CRY2 encoding gene fragment was conducted, allowing for easier detection and purification of the CRY2 protein in subsequent stages.
Human cryptochrome-2 (CRY2) is a core component of the circadian clock, regulating the sleep-wake cycle and biological rhythms. CRY2, alongside CRY1, interacts with the CLOCK-BMAL1 complex, influencing transcriptional repression during the circadian cycle. In chronobiology, CRY2 is pivotal for maintaining circadian rhythms and synchronizing internal processes with environmental cues. Research on CRY2 extends to metabolic disorders, as it affects energy metabolism and glucose homeostasis. Additionally, CRY2 is involved in DNA repair and cancer biology, influencing tumor suppression mechanisms. Studying CRY2 provides insights into circadian regulation, metabolism, and cancer pathways, offering potential applications in treating circadian-related disorders, metabolic diseases, and cancer therapeutics.