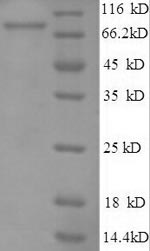
Purity
Greater than 90% as determined by SDS-PAGE.
Research Area
Epigenetics and Nuclear Signaling
Alternative Names
2810024C04Rik; DNA repair protein rad18; E3 ubiquitin-protein ligase RAD18; EC 6.3.2.-; hHR 18; hHR18; hRAD 18; hRAD18; MGC156682; Postreplication repair E3 ubiquitin-protein ligase RAD18; Postreplication repair protein hRAD18p; Postreplication repair protein RAD18; RAD 18; RAD18; RAD18 homolog (S. cerevisiae); RAD18 homolog; RAD18 S. cerevisiae homolog; RAD18 S. cerevisiae homolog of; RAD18 transcript variant; RAD18_HUMAN; Rad18sc; Radiation sensitivity protein 18; RING finger protein 73; RNF 73; RNF73; Structural maintenance of chromosomes protein 6; YCR066W; YCR66W
Species
Homo sapiens (Human)
Expression Region
1-495aa
Target Protein
Sequence
MDSLAESRWPPGLAVMKTIDDLLRCGICFEYFNIAMIIPQCSHNYCSLCIRKFLSYKTQCPTCCVTVTEPDLKNNRILDELVKSLNFARNHLLQFALESPAKSPASSSSKNLAVKVYTPVASRQSLKQGSRLMDNFLIREMSGSTSELLIKENKSKFSPQKEASPAAKTKETRSVEEIAPDPSEAKRPEPPSTSTLKQVTKVDCPVCGVNIPESHINKHLDSCLSREEKKESLRSSVHKRKPLPKTVYNLLSDRDLKKKLKEHGLSIQGNKQQLIKRHQEFVHMYNAQCDALHPKSAAEIVREIENIEKTRMRLEASKLNESVMVFTKDQTEKEIDEIHSKYRKKHKSEFQLLVDQARKGYKKIAGMSQKTVTITKEDESTEKLSSVCMGQEDNMTSVTNHFSQSKLDSPEELEPDREEDSSSCIDIQEVLSSSESDSCNSSSSDIIRDLLEEEEAWEASHKNDLQDTEISPRQNRRTRAAESAEIEPRNKRNRN
Note: The complete
sequence may include tag sequence, target protein sequence, linker sequence and extra sequence that is
translated with the protein sequence for the purpose(s) of secretion, stability, solubility, etc.
If the exact amino acid sequence of this recombinant protein is critical to your application,
please explicitly request the full and complete sequence of this protein before ordering.
Protein Length
Full Length
Tag Info
N-terminal 6xHis-SUMO-tagged
Form
Liquid or Lyophilized powder
Note: We will preferentially ship the format that
we have in stock, however, if you have any special requirement for the format, please remark your
requirement when placing the order, we will prepare according to your demand.
Buffer
If the delivery form is liquid, the default storage buffer is Tris/PBS-based buffer, 5%-50% glycerol.
Note: If you have any special requirement for the
glycerol content, please remark when you place the order.
If the delivery form is lyophilized powder, the buffer before lyophilization is Tris/PBS-based buffer,
6% Trehalose.
Reconstitution
We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20°C/-80°C. Our default final concentration of glycerol is 50%. Customers could use it as reference.
Storage Condition
Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid
repeated freeze-thaw
cycles.
Shelf Life
The shelf life is related to many factors, storage state, buffer ingredients, storage
temperature
and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized
form is 12 months at -20°C/-80°C.
Lead Time
Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
Notes
Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
Datasheet & COA
Please contact us to get it.