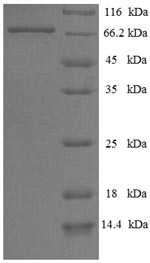
The recombinant Human PRKN was expressed with the amino acid range of 1-465. This PRKN protein is expected to have a theoretical molecular weight of 67.6 kDa. This PRKN recombinant protein is manufactured in e.coli. The PRKN coding gene included the N-terminal 6xHis-SUMO tag, which simplifies the detection and purification processes of the recombinant PRKN protein in following stages of expression and purification.
Parkin (PRKN) is an E3 ubiquitin-protein ligase that plays a crucial role in the regulation of protein degradation through the ubiquitin-proteasome system. It is primarily known for its involvement in maintaining mitochondrial homeostasis and mitophagy, the selective removal of damaged or dysfunctional mitochondria. PRKN helps identify and ubiquitinate proteins on the surface of damaged mitochondria, marking them for degradation. This process is vital for cellular quality control, preventing the accumulation of defective mitochondria and maintaining cellular health. Research areas involving PRKN include neurodegenerative diseases, particularly Parkinson's disease, as mutations in the PRKN gene are associated with familial forms of the disorder. Understanding Parkin's function and dysregulation in disease conditions may provide insights into therapeutic strategies for neurodegenerative disorders and contribute to broader knowledge of cellular quality control mechanisms.