What Is Protein Degradation?
Protein degradation, also known as proteolysis, refers to a set of enzymatic reactions by which the proteins are broken down into smaller polypeptides or amino acids by protease. Proteolysis may also occur in some special conditions that are absent from the enzymes, such as mineral acids and high temperatures. Proteolysis includes two main pathways: ubiquitin-mediated proteolysis and lysosomal proteolysis.
What Is Ubiquitin Mediated Proteolysis?
Ubiquitin mediated proteolysis is the process by which ubiquitin binds covalently to the target protein and degrades the target protein. The ubiquitination and subsequent degradation process of target proteins run through the plasma membrane system of the entire cell, from the cell membrane to the endoplasmic reticulum to the nuclear membrane.
The Function of Ubiquitin Mediated Proteolysis
The ubiquitin-proteasome pathway is an important protein quality control system in eukaryotic cells. Proteolysis by ubiquitin-proteasome pathway regulates the cell cycle, cell proliferation & differentiation, signal transduction, and other cellular physiological processes. And intracellular ubiquitin-mediated proteolysis is one of the most important post-transcriptional modifications of proteins.
The Process of Ubiquitin Mediated Proteolysis
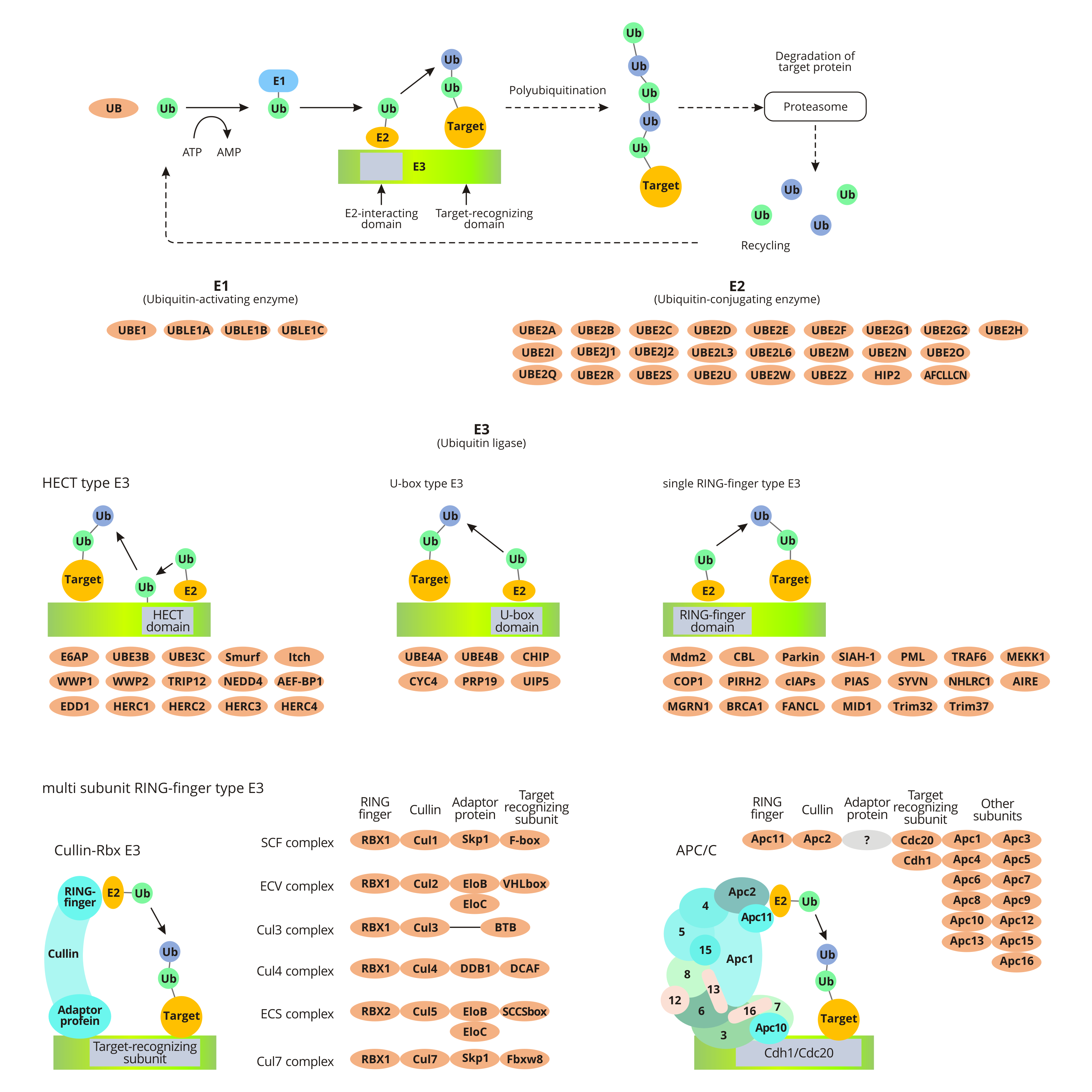
In the pathway of ubiquitin-mediated proteolysis, the substrate protein binds to ubiquitin by ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin ligase E3. Ubiquitin, a small protein, functions as a marker for marking the substrate for degradation by the proteasome (26s proteasome).
Ubiquitination of the target Protein
The ubiquitination of target proteins is a consuming-ATP process. First, E1 catalyzes the adenylation of ubiquitin's C-terminal. Activated ubiquitin transfers to the Cys residues of E1 and subsequently get molecular rearrangement to form intramolecular thioester bonds. Secondly, the E1-ubiquitin complex transfers ubiquitin to the E2 by transacylation. Finally, the ubiquitin ligase E3 binds to the target protein and specifically recognizes E2. And then the target protein binds to the ubiquitin linked to the E2 enzyme. The process means the ubiquitination of the target protein is finished.
When the first ubiquitin molecule is linked to the target protein under the catalysis of E3, other ubiquitin molecules are also successively linked to the Lys residue of the previous ubiquitin molecule to form a polyubiquitin chain. The formation of polyubiquitin chains is a signal for protein degradation. And studies have found that a novel ubiquitin chain transfer factor E4 (Ufd2p) can convert non-degradable ubiquitin chain signals into degraded ubiquitin chain signals. Degraded ubiquitin chain signals can be recognized by the proteasome, which promotes the degradation of modified substrate proteins.
Degradation of the Ubiquitin-protein Conjugate
The 26s proteasome consists of two circular 19s subunits and one microsomal 20s subunit. After the ubiquitin-protein conjugate interacts with the 19s ubiquitin receptor of the 26s proteasome, the protein substrate is unfolded. The unfolded protein substrate enters the 20s core particle through the proteasome receptor end crevice, where the substrate protein is cleaved multiple times and forms a peptide of 3-22 amino acid residues. Under the action of ubiquitin and oligopeptidase, the polyubiquitin chain is reduced to monomer for the next round of protein degradation.
Abnormal Ubiquitin Mediated Proteolysis and Diseases
Because proteins associated with diseases are commonly used as substrates in the ubiquitin degradation pathway. And abnormal ubiquitin-mediated proteolysis could cause an increased or reduced rate of degradation. Therefore, if the ubiquitin metabolic pathway is abnormal, it will lead to a variety of diseases.
Malignant tumors are often related to the expression of oncogenes and tumor suppressor gene products. When growth factors that cause tumors fail to degrade in the ubiquitin-proteasome system, it normally triggers tumors.
For the nervous system, alterations in its substrate homeostasis cause a significant decrease in the excited excitatory junction potential. Studies on Drosophila neuromuscular junctions have shown that overexpression of the faf gene (in the ubiquitinase family) in the ubiquitin system leads to synaptic overgrowth and neurotransmitter release disorders. Angelman syndrome is caused by a lack of a gene encoding E6-AP or an abnormal function of E6-AP protease.





