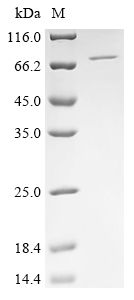
The full-length sequence of the human PARP2 gene (1-583aa) is tagged with 6xHis at the N-terminus to form the target gene. This target gene is amplified using PCR and inserted into a baculovirus transfer vector. The constructed vector is transfected into insect cells such as Sf9 cells, producing recombinant baculovirus. The recombinant baculovirus is collected from the culture medium and further amplified by infecting new insect cells. The amplified recombinant baculovirus transfects new insect cells, which are induced to express the target proteins. The collected proteins are purified by affinity chromatography to obtain the recombinant human PARP2 proteins. The purity of the recombinant PARP2 protein is over 90% as measured by SDS-PAGE.
Human PARP2 plays a crucial role in cellular processes related to DNA repair, genomic stability, and cellular stress responses. Structurally, PARP2 consists of several domains that contribute to its function, including a catalytic domain responsible for its enzymatic activity and a DNA-binding domain that facilitates its interaction with damaged DNA [1]. The crystal structure of PARP2 has revealed insights into its catalytic mechanism and how it interacts with inhibitors, which is particularly relevant for the development of cancer therapies targeting PARP enzymes [2]. Notably, PARP2 is less active than PARP1, which may be attributed to specific structural features within its catalytic domain that influence substrate recognition and binding [3].
PARP2, along with PARP1, is primarily involved in the repair of single-strand breaks (SSBs) in DNA through base excision repair (BER) [4][5]. Compared to PARP1, which exhibits a higher affinity for DNA damage sites, PARP2 has been shown to have a lower activity level in catalyzing ADP-ribosylation of proteins [3][4]. The enzymatic activity of PARP2 is characterized by its ability to transfer ADP-ribose units from NAD+ to target proteins, forming long-branched poly(ADP-ribose) (PAR) polymers that play a role in recruiting other DNA repair proteins to sites of damage [6].
References:
[1] T. Karlberg, M. Hammarstrom, P. Schutz, L. Svensson, & H. Schüler, Crystal structure of the catalytic domain of human parp2 in complex with parp inhibitor abt-888, Biochemistry, vol. 49, no. 6, p. 1056-1058, 2010. https://doi.org/10.1021/bi902079y
[2] M. Aoyagi-Scharber, A. Gardberg, B. Yip, B. Wang, Y. Shen, & P. Fitzpatrick, Structural basis for the inhibition of poly(adp-ribose) polymerases 1 and 2 by bmn 673, a potent inhibitor derived from dihydropyridophthalazinone, Acta Crystallographica Section F Structural Biology Communications, vol. 70, no. 9, p. 1143-1149, 2014. https://doi.org/10.1107/s2053230x14015088
[3] M. Kutuzov, J. Amé, S. Khodyreva, V. Schreiber, & O. Lavrik, Interaction of parp2 with dna structures mimicking dna repair intermediates, Biopolymers and Cell, vol. 27, no. 5, p. 383-386, 2011. https://doi.org/10.7124/bc.000129
[4] M. Kutuzov, E. Belousova, T. Kurgina, A. Ukraintsev, И. Васильева, С. Ходырева, et al., The contribution of parp1, parp2 and poly(adp-ribosyl)ation to base excision repair in the nucleosomal context, Scientific Reports, vol. 11, no. 1, 2021. https://doi.org/10.1038/s41598-021-84351-1
[5] M. Sukhanova, S. Abrakhi, V. Joshi, D. Pastré, M. Kutuzov, R. Anarbaev, et al., Single molecule detection of parp1 and parp2 interaction with dna strand breaks and their poly(adp-ribosyl)ation using high-resolution afm imaging, Nucleic Acids Research, vol. 44, no. 6, p. e60-e60, 2015. https://doi.org/10.1093/nar/gkv1476
[6] I. Talhaoui, Н. Лебедева, G. Zarkovic, C. Saint-Pierre, M. Kutuzov, M. Sukhanova, et al., Poly(adp-ribose) polymerases covalently modify strand break termini in dna fragmentsin vitro, Nucleic Acids Research, p. gkw675, 2016. https://doi.org/10.1093/nar/gkw675