Disease and disorder research has been conducted in relation to the histone modifications pathway and malignant neoplasms, carcinogenesis, cell transformation, neoplastic, carcinoma. The histone modifications pathway has been researched in relation to methylation, DNA methylation, histone acetylation, chromatin remodeling, and histone methylation.
Histone modifications and DNA methylation are epigenetic marks that effect gene expression patterns. As shown in the figure 1, chromosomal DNA is wrapped around histones to form nucleosomes. Histones can be modified by a large number of post-translational modifications (PTMs), especially on histone tails, which signal and result in specific gene expression or repression patterns. There can be interplay between certain histone modifications and DNA methylation, allowing further levels of mechanisms to control and regulate gene expression.
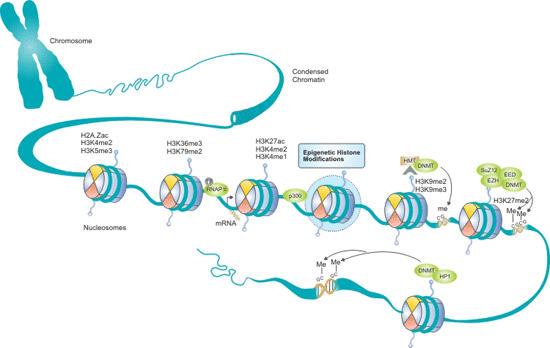
Figure 1. The Interplay between Histone Modifications and DNA Methylation
There are 5 histone families (H1-H5) divided into two groups: core histones (H2A, H2B, H3, and H4) and linker histones (H1 and H5). Post-translational modifications (PTMs) mentioned in the earlier include acetylation, methylation, phosphorylation, and ubiquitination. These modifications function to regulate both nucleosome structure and stability, and the recruitment of chromatin binding proteins. Here, we summarize two histone modifications interactive pathway, H2A, H2B, and H4 Interactive pathway and H3 interactive pathway.
1. H2A, H2B, and H4 Interactive pathway
Generally speaking, based on different function, there are three types of enzymes in the process of histone modification and interacting with histones to affect chromatin structure and transcription, involving writers, readers and erasers. Writers are enzymes that add PTMs, while erasers are enzymes that remove PTMs. Reader proteins bind to the PTMs and function to regulate changes in chromatin structure and gene expression.
This interactive pathway discusses the writers and erasers for histones H2A, H2B and histone H4, as well as the amino acid residues they modify. As the Figure 2 shows, Histone acetyltransferases (HATs) are writers that acetylate lysine residues, including histone H2A Lys5, H2B Lys5, 12, 15, 20, and H4 Lys5, 8, 12, 16. Conversely, the deacytelation of these residues is performed by erasers known as histone deacetylases (HDACs). Acetylation of lysine residues makes chromatin accessible to transcription factors and can significantly increase gene expression.
Then, Histone lysine methyltransferases (KMTs) are writers that add methyl groups to lysine residues, which may be removed by erasers known as histone lysine demethylases (KDMs). Lysines can be either mono-, di-, or tri-methylated, forming functional diversity to each site of methylation.
Additionally, multiple kinases and phosphatases can phosphorylate and dephosphorylate serine, threonine, and tyrosine residues on histone proteins, respectively. The best-known function of histone phosphorylation takes place during cellular response to DNA damage, when phosphorylated histone H2A(X) demarcates large chromatin domains around the site of DNA breakage.
Lastly, histone ubiquitination occurs when small, 76-amino acid ubiquitin molecules are attached to lysine residues with 3 specialized enzymes called the E1-activating, E2-conjugating, and E3-ligase enzymes. If you want to know more thing of histone modifications or related antibody products, you can click the article entitled Four Common Histone Modifications".
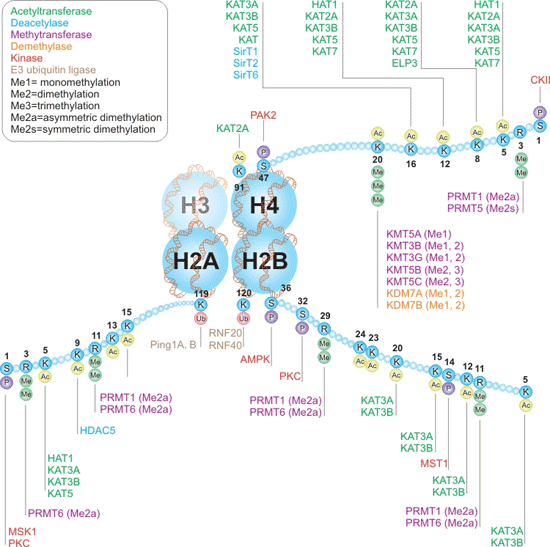
Figure 2. Writers and Erasers of Histones H2A, H2B, and H4 Interactive Pathway
2. H3 interactive pathway
Histone H3 is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells, and also plays a critical roles in the regulation of chromatin structure and gene expression. Compared with other four types of histones, the modifications of histone H3 just involve acetylation, methylation, and phosphorylation. These PTMs can occur at different residues on histone H3 and regulate various processes involving nuclear organization, chromatin structure, and recruitment of chromatin binding proteins (also called reader proteins), as the Figure 2 shows.
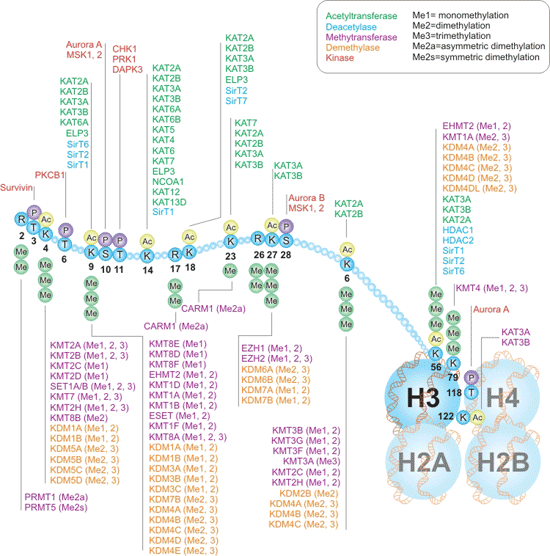
Figure 3. Writers and Erasers of Histones H3 Interactive Pathway
CUSABIO team. Epigenetic Histone Modifications Pathway. https://www.cusabio.com/c-20825.html







Comments
Leave a Comment