Alternative Names
Eukaryotic peptide chain release factor subunit 1 (Eukaryotic release factor 1) (eRF1) (Omnipotent suppressor protein 1) SUP45 SAL4 SUP1 YBR143C YBR1120
Species Reactivity
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
Immunogen
Recombinant Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) SUP45 protein (26-262aa)
Immunogen Species
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
Purification Method
Affinity-chromatography
Concentration
It differs from different batches. Please contact us to confirm it.
Buffer
Preservative: 0.03% Proclin 300
Constituents: 50% Glycerol, 0.01M PBS, pH 7.4
Tested Applications
ELISA, WB
Storage
Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
Lead Time
Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
Description
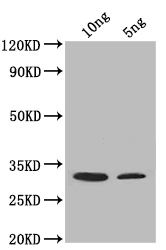
The procedure for generating polyclonal antibodies against SUP45 involves repetitive immunizations of a rabbit with recombinant Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) SUP45 protein (26-262aa). Upon reaching a sufficient antibody titer, the rabbit undergoes bleeding, and the serum-derived antibodies are purified through affinity chromatography. Two applications ELISA and WB have been used in assays to test for the specificity of this SUP45 antibody. The SUP45 antibody shows specific reactivity towards Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) SUP45 protein.
The SUP45 is a vital component of the translation termination machinery in yeast. Its primary function lies in facilitating the accurate and timely termination of protein synthesis. Acting as a translation release factor, SUP45 catalyzes the hydrolysis of the ester bond between the polypeptide chain and the tRNA molecule, ensuring the proper release of the newly synthesized protein from the ribosome when a stop codon is encountered. SUP45 also contributes to quality control mechanisms by recognizing and responding to premature termination events, thereby preventing the synthesis of incomplete or faulty proteins.
Usage
For Research Use Only. Not for use in diagnostic or therapeutic procedures.