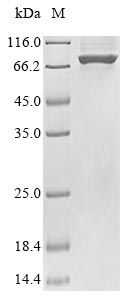
The gene fragment corresponding to the 1-720aa of the E.coli uvrD protein was cloned into an expression vector. The expression vector was transformed into the E.coli for expression. The generated product was purified and separated to obtain the recombinant E.coli uvrD protein. Its purity is higher than 85% measured by SDS-PAGE. This recombinant uvrD protein showed an apparent molecular weight of about 90 kDa under SDS-PAGE condition.
uvrD is a gene encoding a protein named DNA helicase II and belongs helicase family and UvrD subfamily. Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genes.uvrD is a helicase with DNA-dependent ATPase activity and has a critical role in the processing of DNA-protein cross-links. It is involved in the post-incision events of nucleotide excision repair and methyl-directed mismatch repair, and probably also in repair of alkylated DNA.