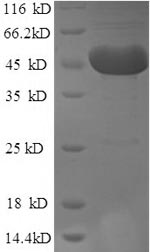
Producing recombinant human m7GpppN-mRNA hydrolase (DCP2) involves a series of steps beginning with the isolation of the target gene that codes for the 1-385aa of the human DCP2, which is also fused with N-terminal 6xHis-tag for purification purposes. This gene is cloned into an expression vector and transfected into E. coli cells. The E. coli cells express the protein, which is subsequently harvested from the cell lysate. Purification of the protein is commonly achieved using affinity chromatography. Finally, the recombinant DCP2 protein's purity is determined by SDS-PAGE, reaching over 90%.
Human DCP2 is a catalytically active mRNA-decapping enzyme found in specific cytoplasmic structures [1]. It is an RNA-binding protein that selectively binds to a subset of mRNAs containing a stem-loop structure near the 5' cap [2]. DCP2 plays a crucial role in controlling the stability of a subset of human cellular RNAs involved in various functions such as transcription and immune responses [3]. The activation of DCP2 by DCP1 occurs on the EDC4 scaffold and is involved in mRNA degradation by XRN1 in human cells [4]. DCP2 cooperates with MOV10 to de-cap LINE-1 RNA, leading to the degradation of LINE-1 RNA and a reduction in LINE-1 retrotransposition [5].
In humans, the DCP2-DCP1 interaction requires additional proteins like EDC3, ECD4, and the DEAD-box protein DDX6/RCK to form multimeric decapping complexes [6]. DCP2 is regulated at the post-transcriptional level, with miRNAs being an effective way to downregulate its expression [7]. Furthermore, DCP2 preferentially binds to specific mRNAs and has been identified to have specific substrates, such as Rrp41, a core subunit component of the RNA exosome [8].
References:
[1] E. Dijk, N. Cougot, S. Meyer, S. Babajko, E. Wahle, & B. Séraphin, Human dcp2: a catalytically active mrna decapping enzyme located in specific cytoplasmic structures, The Embo Journal, vol. 21, no. 24, p. 6915-6924, 2002. https://doi.org/10.1093/emboj/cdf678
[2] E. Grudzien-Nogalska and M. Kiledjian, New insights into decapping enzymes and selective mrna decay, Wiley Interdisciplinary Reviews - Rna, vol. 8, no. 1, 2016. https://doi.org/10.1002/wrna.1379
[3] Y. Luo, Z. Na, & S. Slavoff, Phage-display selection of a cell-permeable bicyclic peptide as a selective ligand of the human rna decapping enzyme dcp2,, 2020. https://doi.org/10.26434/chemrxiv.12233102.v1
[4] C. Chang, N. Bercovich, B. Loh, S. Jonas, & E. Izaurralde, The activation of the decapping enzyme dcp2 by dcp1 occurs on the edc4 scaffold and involves a conserved loop in dcp1, Nucleic Acids Research, vol. 42, no. 8, p. 5217-5233, 2014. https://doi.org/10.1093/nar/gku129
[5] Q. Liu, D. Yi, J. Ding, Y. Mao, S. Wang, L. Maet al., mov10 recruits dcp2 to decap human line‐1 rna by forming large cytoplasmic granules with phase separation properties, Embo Reports, vol. 24, no. 9, 2023. https://doi.org/10.15252/embr.202256512
[6] F. Tritschler, J. Braun, C. Motz, C. Igreja, G. Haas, V. Truffaultet al., Dcp1 forms asymmetric trimers to assemble into active mrna decapping complexes in metazoa, Proceedings of the National Academy of Sciences, vol. 106, no. 51, p. 21591-21596, 2009. https://doi.org/10.1073/pnas.0909871106
[7] Q. Zhang, Y. Yan, Q. Lv, Y. Li, R. Wang, G. Sunet al., Mir-4293 upregulates lncrna wfdc21p by suppressing mrna-decapping enzyme 2 to promote lung carcinoma proliferation, Cell Death and Disease, vol. 12, no. 8, 2021. https://doi.org/10.1038/s41419-021-04021-y
[8] Y. Li, M. Song, & M. Kiledjian, Transcript-specific decapping and regulated stability by the human dcp2 decapping protein, Molecular and Cellular Biology, vol. 28, no. 3, p. 939-948, 2008. https://doi.org/10.1128/mcb.01727-07