MAD2L2 is a chromatin-binding protein that participates in the modulation of cell cycle and DNA damage response (DDR). MAD2L2 is involved in preventing the onset of anaphase and ensuring that all chromosomes are aligned correctly at the metaphase plate. It also takes part in translesion DNA synthesis (TLS), mitotic control, signal transduction, transcription, as well as the choice of repair pathway in DNA double-strand breaks. MAD2L2 was previously described as a non-catalytic auxiliary subunit of DNA pol zeta (ϛ), and its knockdown caused hypersensitivity to DNA damage. MAD2L2 dysregulation has been found in a variety of cancers, and MAD2L2 overexpression has been detected in glioma, epithelial ovarian cancer, and breast cancer.
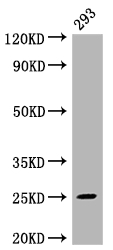
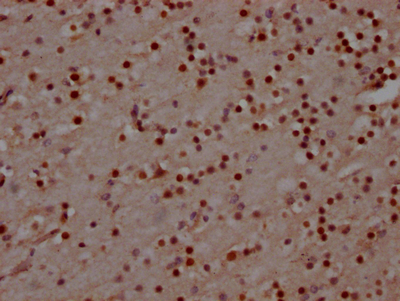
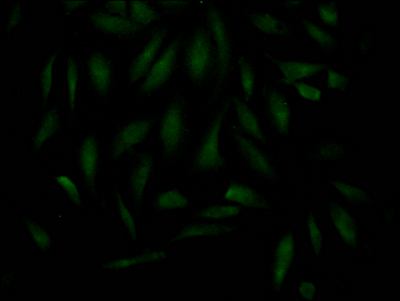
CUSABIO cloned MAD2L2 antibody-coding genes into plasma vectors and then transfected these vector clones into mammalian cells using a lipid-based transfection reagent. Following transient expression, the recombinant antibodies against MAD2L2 were harvested and characterized. The recombinant MAD2L2 antibody was purified by Affinity-chromatography from the culture medium. It can be used to detect MAD2L2 protein from Human in the ELISA, WB, IHC, IF.