Histones are fundamental proteins that play a vital role in the packaging, organization, and regulation of DNA within the nucleus of eukaryotic cells. They form the building blocks of chromatin, the complex structure in which DNA is wrapped, enabling efficient storage and transmission of genetic information.
Understanding the composition, structure, and functions of histones provides invaluable insights into the intricate relationship between these proteins and DNA, shedding light on both normal cellular processes and disease states such as cancer.
In this article, we'll take a closer look at histones. We'll see what histones are, their classification, how the structure of histones supports their function, and how histones cause diseases.
1. What Are Histones?
Histones were initially isolated from avian red blood cells and were described by Albrecht Kossel in 1884. Histones are DNA-binding proteins existing in the chromatin of all eukaryotic cells. Histones contain abundant basic amino acids arginine and lysine that confer their positive charges, allowing for their tight binding with negatively charged DNA, promoting the formation of nucleosomes, and facilitating chromatin compaction.
Histones have a globular core domain and an unstructured N-terminal tail. The core domain facilitates the assembly of histone octamers, while the N-terminal tail extends out from the nucleosome and undergoes various posttranslational modifications (PTMs), including acetylation, methylation, ubiquitylation, phosphorylation, and more. This unstructured region provides flexibility and allows for various modifications that regulate chromatin structure and gene expression [1]. The core domain contains the histone fold, a three-helix bundle that allows for protein-protein interactions, DNA binding, and chromatin compaction [2].
2. Classification of Histones
Histones are mainly divided into five categories: H2A, H2B, H3, and H4 (core histones), and linker histone: H1 / H5 [3]. H2A, H2B, H3, and H4 histones form the core of the nucleosome and actively participate in the precise wrapping of DNA. Linker histone H1 associates with the linker region of DNA between two nucleosomes, facilitating the construction of numerous nucleosomes into higher-order chromatin structures. H1 varies greatly between species. As the variant of H1, H5 is the major linker histone in avian erythrocytes.
Table: Histone family members
| Super family |
Family |
Location and Function |
Subfamily |
Members |
| Linker |
H1 |
On the connecting line; bind to nucleosomes and linker-DNA in the chromatin fiber |
H1F |
H1F0, H1FNT, H1FOO, H1FX |
| H1H1 |
HIST1H1A, HIST1H1B, HIST1H1C, HIST1H1D, HIST1H1E, HIST1H1T |
| Core |
H2A |
Core particles; take part in the formation of nucleosomes |
H2AF |
H2AFB1, H2AFB2, H2AFB3, H2AFJ, H2AFV, H2AFX, H2AFY, H2AFY2, H2AFZ |
| H2A1 |
HIST1H2AA, HIST1H2AB, HIST1H2AC, HIST1H2AD, HIST1H2AE, HIST1H2AG, HIST1H2AI,HIST1H2AJ, HIST1H2AK, HIST1H2AL, HIST1H2AM |
| H2A2 |
HIST2H2AA3, HIST2H2AC |
| H2B |
Core particles; take part in the formation of nucleosomes |
H2BF |
H2BFM, H2BFS, H2BFWT |
| H2B1 |
HIST1H2BA, HIST1H2BB, HIST1H2BC, HIST1H2BD, HIST1H2BE, HIST1H2BF, HIST1H2BG,HIST1H2BH, HIST1H2BI, HIST1H2BJ, HIST1H2BK, HIST1H2BL, HIST1H2BM, HIST1H2BN, HIST1H2BO |
| H2B2 |
HIST2H2BE |
| H3 |
Core particles; take part in the formation of nucleosomes |
H3A1 |
HIST1H3A, HIST1H3B, HIST1H3C, HIST1H3D, HIST1H3E, HIST1H3F, HIST1H3G, HIST1H3H, HIST1H3I, HIST1H3J |
| H3A2 |
HIST2H3C |
| H3A3 |
HIST3H3 |
| H4 |
Core particles; take part in the formation of nucleosomes |
H41 |
HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L |
| H44 |
HIST4H4 |
3. Function of Histones
In the nucleus, histones are primarily responsible for compacting DNA strands and forming nucleosomes, which are the basic building blocks of chromatin [4]. By wrapping DNA around an octamer of histone proteins, histones enable the efficient storage of genetic material and protect it from damage.
Histones are also involved in the regulation of gene expression. Modifications to histones, such as acetylation, methylation, phosphorylation, and ubiquitination, act as an epigenetic code, marking specific regions of the genome for different regulatory processes. They can either promote or inhibit the binding of proteins involved in gene regulation, influencing the accessibility of DNA to transcription factors and other regulatory proteins and affecting gene expression patterns [5].
For example, acetylation of histones is generally associated with open chromatin and active gene transcription, while methylation can have activating or repressive effects depending on the specific amino acid and degree of methylation.
Extensive evidence has illuminated the critical involvement of histone modifications in nearly all DNA-based processes, encompassing gene transcription, DNA replication, DNA damage repair, and DNA recombination [6-10].
Histone modifications contribute to epigenetic regulation. Histone modifications, along with other epigenetic marks such as DNA methylation, are involved in establishing and maintaining these patterns of gene expression.
At the cell surface, histones promote cell-mediated apoptosis, neurogenesis, migration, and endocytosis. Additionally, emerging studies show that histones are released into the extracellular space, where histones exert remarkable toxic or pro-inflammation activity in vivo and in vitro [11][12].
4. Histone-related Diseases
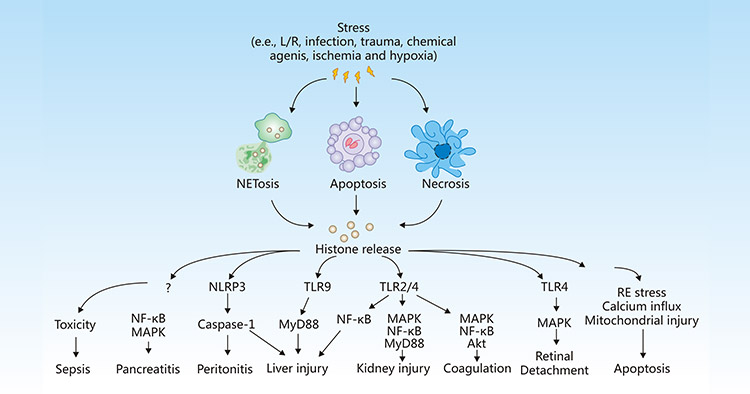
Histones are the basic structural proteins of eukaryotic chromatin. However, under stress conditions such as inflammation, infection, ischemia, and hypoxia, cells may undergo apoptosis, necrosis, or NETosis, thus leading to the release of histones into the extracellular space, where histones can activate immune and inflammatory responses, initiating or aggravating tissue injury, or mediating the occurrence and development of various diseases, including sepsis, pancreatitis, peritonitis, liver injury, kidney injury, coagulation, detachment, and apoptosis.
Figure 1:Release and activity of histones in response to stress
The picture is cited from https://www.nature.com/articles/cddis2014337
During sepsis, pro-inflammatory factors TNF-α and IL-6 are released in large quantities, triggering the efflux of histones into extracellular space, thus resulting in endothelial dysfunction, organ failure, and death [12].
Kang R et al. showed that extracellular histone-mediated HMGB1 release in activated immune cells contributes to L-arginine-induced acute pancreatitis in HMGB1 pancreatic conditional knockout mice [13].
Extracellular histones can activate the NLRP3 inflammasome and indirectly induce cell necrosis to produce local cytokines, thus leading to peritonitis [14].
Once released, histones selectively bind to TLRs, including TLR-2, TLR-4, and TLR9 to generate pro-inflammatory cytokines, which enhance inflammatory response and tissue damage in the liver [15][16], kidney [17], lung [18], and brain. The TLR2 and TLR4 signaling pathways induced also promote platelet activation, blood coagulation, increase the level of von Willebrand factor, and accelerate the early occurrence of deep vein thrombosis [19].
Kawano H et al. revealed that histones released from damaged retinal in retinal detachment (RD) may deteriorate the condition of the subretinal microenvironment by inducing inflammation and cell death in retinal pigment epithelial (RPE) cells [20].
Histones released from NETosis have been involved in numerous autoimmune and autoinflammatory diseases such as systemic lupus, rheumatoid arthritis, and small-vessel vasculitis.
Extracellular histones and histone modifications also correlate with the pathogenesis of neurological diseases, including ischemia-reperfusion injury, histone modification-mediated transcription disorders, and glial cell reactive hyperplasia.
Additionally, mutations in genes encoding histones or their modifying enzymes have been identified in different cancer types, highlighting their significance in tumor development. These mutations can affect the stability of nucleosomes, disrupt the regulation of gene expression, and contribute to oncogenesis. The dysregulated expression of histone modifier enzymes specifically has been strongly linked to the disruption of the histone modification machinery, ultimately resulting in the initiation, progression, and metastasis of cancer.
For example, the transcription factor FOXA1 orchestrates extensive enhancer reprogramming by modulating H3K27ac, consequently facilitating the progression of pancreatic cancer (PC). In hepatocellular carcinoma (HCC), an upregulation of H3K9me2 in the promoter region of the RARRES3 gene suppresses its transcription, thereby promoting cancer cell migration. Histone modifications have been explored as potential diagnostic and prognostic markers in cancer, aiding in personalized treatment strategies.
In summary, histones are the enigmatic guardians of DNA, orchestrating its packaging, regulation, and transmission. Their composition, structure, and modifications allow for precise control of chromatin structure and gene expression. Understanding the intricate relationship between histones and DNA provides valuable insights into both normal cellular processes and disease states, particularly cancer. Further exploration of histone biology promises to uncover novel therapeutic targets and advance personalized treatment approaches, offering hope for improved outcomes for patients with cancer and other diseases.
References
[1] Peterson CL, Laniel MA. Histories and histone modifications [J]. Curr Biol. 2004;14(14):R546–R551.
[2] Arents G, Moudrianakis EN. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization [J]. Proc Natl Acad Sci USA. 1995;92(24):11170–11174.
[3] Bhasin M, Reinherz EL, et al. Recognition and classification of histones using support vector machine [J]. J Comput Biol. 2006 Jan-Feb;13(1):102-12.
[4] Campos EI, Reinberg D. Histones: annotating chromatin [J]. Annu Rev Genet 2009; 43: 559–599.
[5] Carlberg C., Molnár F. The Histone Code. In: Human Epigenomics. Springer, Singapore, 2018.
[6] Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression [J]. Annu. Rev. Biochem 2006, 75, 243–269.
[7] Li B, Carey M, et al. The role of chromatin during transcription [J]. Cell 2007, 128, 707–719.
[8] Unnikrishnan A, Gafken PR, et al. Dynamic changes in histone acetylation regulate origins of DNA replication [J]. Nat. Struct. Mol. Biol 2010, 17, 430–437.
[9] Jacob Y, Stroud H, et al. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases [J]. Nature 2010, 466, 987–991.
[10] Scully R. A histone code for DNA repair [J]. Nat. Rev. Mol. Cell. Biol 2010, 11, 164.
[11] Allam R, Kumar SV, et al. Extracellular histones in tissue injury and inflammation [J]. J Mol Med (Berl) 2014; 92: 465–472.
[12] Xu J, Zhang X, et al. Extracellular histones are major mediators of death in sepsis [J]. Nat Med 2009; 15: 1318–1321.
[13] Kang R, Zhang Q, et al. Intracellular Hmgb1 Inhibits Inflammatory Nucleosome Release and Limits Acute Pancreatitis in Mice [J]. Gastroenterology 2014; 146: 1097–1107.
[14] Allam, R., Darisipudi, M. N., et al. Histones trigger sterile inflammation by activating the NLRP3 inflammasome [J]. Eur. J. Immunol. 2013, 43, 3336–3342.
[15] Huang H, Evankovich J, et al. Endogenous histones function as alarmins in sterile inflammatory liver injury through Toll-like receptor 9 in mice [J]. Hepatology 2011; 54: 999–1008.
[16] Wen Z, Liu Y, et al. Circulating histones exacerbate inflammation in mice with acute liver failure [J]. J Cell Biochem 2013; 114: 2384–2391.
[17] Allam R, Scherbaum CR, et al. Histones from dying renal cells aggravate kidney injury via TLR2 and TLR4[J]. J Am Soc Nephrol 2012; 23: 1375–1388.
[18] Bosmann M, Grailer JJ, et al. Extracellular histones are essential effectors of C5aR-and C5L2-mediated tissue damage and inflammation in acute lung injury[J]. Faseb J 2013; 27: 5010–5021.
[19] Carestia A, Rivadeneyra L, et al. Functional responses and molecular mechanisms involved in histone-mediated platelet activation [J]. Thromb Haemost 2013; 110: 1035–1045.
[20] Kawano H, Ito T, et al. Toxic effects of extracellular histones and their neutralization by vitreous in retinal detachment [J] Lab Invest 2014; 94: 569–585.
CUSABIO team. What Are Histones: Unraveling Their Critical Roles in Disease Pathogenesis. https://www.cusabio.com/c-21029.html




Comments
Leave a Comment