Full Product Name
Rabbit anti-Escherichia coli (strain K12) rpoD Polyclonal antibody
Alternative Names
rpoD antibody; alt antibody; b3067 antibody; JW3039 antibody; RNA polymerase sigma factor RpoD antibody; Sigma-70 antibody
Species Reactivity
Escherichia coli (strain K12)
Immunogen
Recombinant Escherichia coli (strain K12) RPOD protein
Immunogen Species
Escherichia coli (strain K12)
Purification Method
Protein A/G
Concentration
It differs from different batches. Please contact us to confirm it.
Buffer
Preservative: 0.03% Proclin 300
Constituents: 50% Glycerol, 0.01M PBS, pH 7.4
Tested Applications
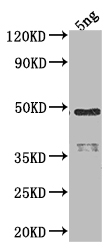
ELISA, WB
Storage
Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
Lead Time
Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
Description
The RPOD antibody is created by immunizing a rabbit with a recombinant RPOD protein derived from Escherichia coli (strain K12). It is purified using protein A/G affinity chromatography. This specific RPOD antibody has the capability to detect the RPOD protein of Escherichia coli (strain K12) in ELISA and WB applications.
The Escherichia coli (strain K12) RPOD protein, also known as the sigma factor σ70, mainly functions to initiate the transcription process in bacteria. RPOD protein binds to the RNA polymerase enzyme and guides it to promoters. By recognizing and binding to these promoters, RPOD helps initiate the transcription of genes into mRNA, which is then used as a template for protein synthesis. RPOD is responsible for the transcription of a large number of genes involved in various cellular processes in E. coli.
Usage
For Research Use Only. Not for use in diagnostic or therapeutic procedures.