Alternative Names
FACT complex subunit SPT16 (Cell division control protein 68) (Facilitates chromatin transcription complex subunit SPT16) (Suppressor of Ty protein 16) SPT16 CDC68 SSF1 YGL207W
Species Reactivity
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
Immunogen
Recombinant Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) SPT16 protein (647-964aa)
Immunogen Species
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
Purification Method
Affinity-chromatography
Concentration
It differs from different batches. Please contact us to confirm it.
Buffer
Preservative: 0.03% Proclin 300
Constituents: 50% Glycerol, 0.01M PBS, pH 7.4
Tested Applications
ELISA, WB
Storage
Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
Lead Time
Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
Description
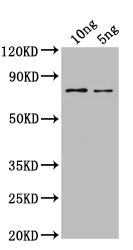
The SPT16 polyclonal antibody is produced by immunizing a rabbit with a recombinant Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) SPT16 protein (647-964aa). The rabbit will mount a humoral response to the immunogen and the antibodies produced can be harvested by bleeding the rabbit to obtain IgG-rich serum. The resulting SPT16 antibody is subjected to affinity chromatography purification. It shows reactivity with Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) SPT16 protein. The functionality of this SPT16 antibody has been validated in ELISA and WB applications.
SPT16 is a component of the FACT complex, which is involved in the reorganization of nucleosomes during transcription. This protein plays a crucial role in maintaining proper chromatin structure, facilitating transcription initiation and elongation, and ensuring accurate gene expression. SPT16 helps prevent the stalling of RNA polymerase and aids in the smooth progression of the transcription process, ultimately contributing to gene regulation and efficient transcription in yeast cells.
Usage
For Research Use Only. Not for use in diagnostic or therapeutic procedures.