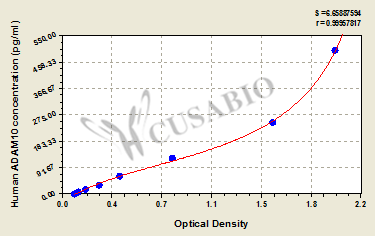
The ELISA Kit is designed for quantitatively measuring human ADAM10 levels in samples, including serum, plasma, or tissue homogenates. It uses the sandwich enzyme immunoassay technique in combination with the enzyme-substrate chromogenic reaction to quantify the analyte in the sample. The color develops positively to the amount of ADAM10 in samples. The color intensity is measured at 450 nm via a microplate reader.
ADAM10, a member of the large family of ADAMs, is expressed at high levels in the brain and modulates the molecular organization and activity of the excitatory synapse by shedding postsynaptic proteins, including N-cadherin (N-CAD), amyloid precursor protein, nectin-1, prion protein, neuroligin-1, neural cell adhesion molecule L1, and ephrin A2 and A5. ADAM10-mediated shedding of cell-adhesion molecules at the synapse activates a number of processes critical to the proper formation, maintenance, and function of the excitatory synaptic circuitries. ADAM10 is essential for development because it cleaves Notch proteins to induce Notch signaling and regulate cell fate decisions. Impairments in ADAM10 level and/or activity are detrimental to the human brain. ADAM10 has been associated with epilepsy, Alzheimer's disease, and the developmental disorder Fragile X syndrome.