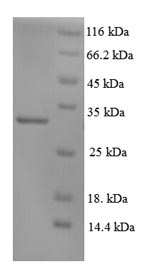
The recombinant Human XPC was expressed with the amino acid range of 496-734. The calculated molecular weight for this XPC protein is 31.5 kDa. This XPC protein is produced using e.coli expression system. The N-terminal 6xHis tag was smoothly integrated into the coding gene of XPC, which enables a simple process of detecting and purifying the XPC recombinant protein in the following steps.
The human DNA repair protein complementing XP-C cells (XPC) is a key component of the nucleotide excision repair (NER) pathway, which is responsible for identifying and repairing DNA damage caused by ultraviolet (UV) light, environmental carcinogens, and other sources. XPC specifically recognizes and binds to DNA lesions, such as bulky adducts and thymidine dimers, marking them for subsequent repair. This initial recognition step is crucial for the recruitment of other NER factors and the removal of damaged DNA segments. Dysfunction or mutations in the XPC gene can lead to xeroderma pigmentosum (XP), a rare genetic disorder characterized by extreme sensitivity to UV radiation and a high risk of skin cancer. The study of XPC is essential for understanding the molecular mechanisms underlying DNA repair processes and how defects in these mechanisms contribute to human diseases, particularly in the context of cancer susceptibility and genomic stability.