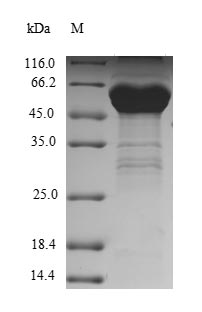
Very nice to receive your inquiry. CSB-YP021801HU
1) This is a developed protein. The product details are confirmed as below.
Code: CSB-YP021801HU
Name: Recombinant Human Transcription activator BRG1(SMARCA4),partial
Expression Region: 700-1246aa; Partial
Tag Info: N-terminal 6xHis-tagged
Expression Sequence:
EVDARHIIENAKQDVDDEYGVSQALARGLQSYYAVAHAVTERVDKQSALMVNGVLKQYQIKGLEWLVSLYNNNLNGILADEMGLGKTIQTIALITYLMEHKRINGPFLIIVPLSTLSNWAYEFDKWAPSVVKVSYKGSPAARRAFVPQLRSGKFNVLLTTYEYIIKDKHILAKIRWKYMIVDEGHRMKNHHCKLTQVLNTHYVAPRRLLLTGTPLQNKLPELWALLNFLLPTIFKSCSTFEQWFNAPFAMTGEKVDLNEEETILIIRRLHKVLRPFLLRRLKKEVEAQLPEKVEYVIKCDMSALQRVLYRHMQAKGVLLTDGSEKDKKGKGGTKTLMNTIMQLRKICNHPYMFQHIEESFSEHLGFTGGIVQGLDLYRASGKFELLDRILPKLRATNHKVLLFCQMTSLMTIMEDYFAYRGFKYLRLDGTTKAEDRGMLLKTFNEPGSEYFIFLLSTRAGGLGLNLQSADTVIIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKILAAAKYKLNVDQKVIQAGMFDQKSSSHERRAF
Product Type: Developed Recombinant Protein
2) The MW of the N-terminal 6xHis-tag is 2.0 kDa. The tag sequence is as below: EAEAYVHHHHHHEFRT
3) This protein was prepared without insertion of restriction sites and was unable to remove the tag.
4) The full-length protein is 1590aa and the MW is ~180 kDa. The above protein we provide is a fragment with 700-1216aa expression region and the MW is ~62.8 kDa. The tag sequence is as below.
EVDARHIIENAKQDVDDEYGVSQALARGLQSYYAVAHAVTERVDKQSALMVNGVLKQYQIKGLEWLVSLYNNNLNGILADEMGLGKTIQTIALITYLMEHKRINGPFLIIVPLSTLSNWAYEFDKWAPSVVKVSYKGSPAARRAFVPQLRSGKFNVLLTTYEYIIKDKHILAKIRWKYMIVDEGHRMKNHHCKLTQVLNTHYVAPRRLLLTGTPLQNKLPELWALLNFLLPTIFKSCSTFEQWFNAPFAMTGEKVDLNEEETILIIRRLHKVLRPFLLRRLKKEVEAQLPEKVEYVIKCDMSALQRVLYRHMQAKGVLLTDGSEKDKKGKGGTKTLMNTIMQLRKICNHPYMFQHIEESFSEHLGFTGGIVQGLDLYRASGKFELLDRILPKLRATNHKVLLFCQMTSLMTIMEDYFAYRGFKYLRLDGTTKAEDRGMLLKTFNEPGSEYFIFLLSTRAGGLGLNLQSADTVIIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKILAAAKYKLNVDQKVIQAGMFDQKSSSHERRAF
So the MW of the fusion protein is 64.8 kDa (2+62.8=64.8).
5) The tags successfully expressed for the MP expression system include N-terminal 10xHis-tagged and C-terminal Myc-tagged, N-terminal 6xHis-tagged, N-terminal 10xHis-tagged, N-terminal Flag-Myc-tagged,
C-terminal FC-tagged, etc.
CSB-MP021801HU >>Mammalian cell
Expression Region: 700-1246aa; Partial
Expression Sequence:
EVDARHIIENAKQDVDDEYGVSQALARGLQSYYAVAHAVTERVDKQSALMVNGVLKQYQIKGLEWLVSLYNNNLNGILADEMGLGKTIQTIALITYLMEHKRINGPFLIIVPLSTLSNWAYEFDKWAPSVVKVSYKGSPAARRAFVPQLRSGKFNVLLTTYEYIIKDKHILAKIRWKYMIVDEGHRMKNHHCKLTQVLNTHYVAPRRLLLTGTPLQNKLPELWALLNFLLPTIFKSCSTFEQWFNAPFAMTGEKVDLNEEETILIIRRLHKVLRPFLLRRLKKEVEAQLPEKVEYVIKCDMSALQRVLYRHMQAKGVLLTDGSEKDKKGKGGTKTLMNTIMQLRKICNHPYMFQHIEESFSEHLGFTGGIVQGLDLYRASGKFELLDRILPKLRATNHKVLLFCQMTSLMTIMEDYFAYRGFKYLRLDGTTKAEDRGMLLKTFNEPGSEYFIFLLSTRAGGLGLNLQSADTVIIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTVNSVEEKILAAAKYKLNVDQKVIQAGMFDQKSSSHERRAF
Here we provide another expression region for your reference.
CSB-MP021801HU1 >>Mammalian cell
Expression Region: 808-1092aa; Partial
Tag Type: The tag type will be determined during the production process.
Expression Region:
IIVPLSTLSNWAYEFDKWAPSVVKVSYKGSPAARRAFVPQLRSGKFNVLLTTYEYIIKDKHILAKIRWKYMIVDEGHRMKNHHCKLTQVLNTHYVAPRRLLLTGTPLQNKLPELWALLNFLLPTIFKSCSTFEQWFNAPFAMTGEKVDLNEEETILIIRRLHKVLRPFLLRRLKKEVEAQLPEKVEYVIKCDMSALQRVLYRHMQAKGVLLTDGSEKDKKGKGGTKTLMNTIMQLRKICNHPYMFQHIEESFSEHLGFTGGIVQGLDLYRASGKFELLDRILPKL