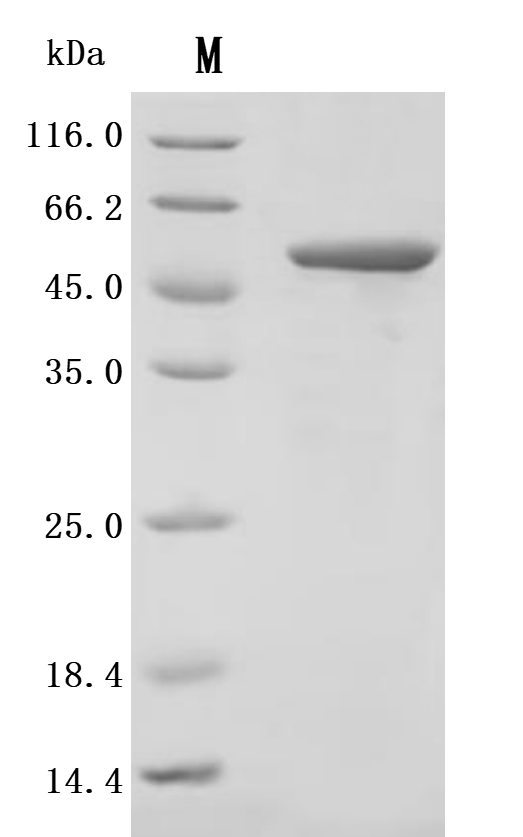
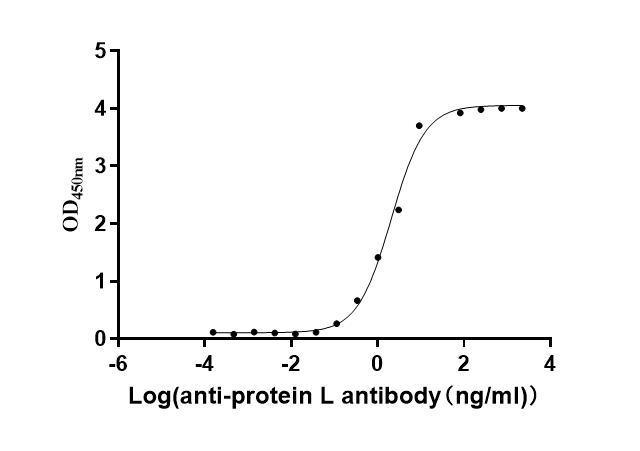
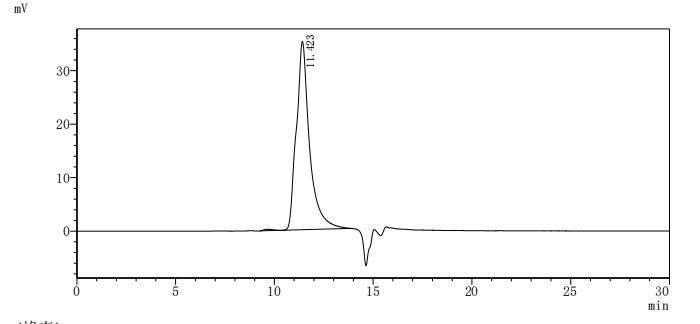
The recombinant LPXTG-motif cell wall anchor domain protein from Finegoldia magna ATCC 53516 is an active protein expressed in a mammalian system to ensure proper folding and functional integrity. This protein covers amino acids 106 to 470 of the native sequence and includes an N-terminal 10xHis tag for convenient purification and detection. Supplied as a lyophilized powder, it demonstrates exceptional purity, greater than 95% as confirmed by both SDS-PAGE and SEC-HPLC. Functional activity was assessed using ELISA, where the recombinant LPXTG-motif cell wall anchor domain protein binds specifically to anti-protein L recombinant antibody (CSB-RA006163MA1HU), with an EC50 ranging from 1.601 to 1.944 ng/mL. These results validate its utility in studies involving bacterial adhesion mechanisms, antibody interaction assays, and microbiome-related research.
The LPXTG-motif cell wall anchor domain protein is significant for Finegoldia magna, a Gram-positive anaerobic bacterium. This protein domain is recognized as essential for attaching proteins to the bacterial cell wall via the action of sortases, which cleave the LPXTG motif and facilitate this anchoring process. Specifically, strain ATCC 53516 of Finegoldia magna may express such proteins. However, detailed molecular characterizations are limited and merit further investigation.
Finegoldia magna is an increasingly recognized opportunistic pathogen, often isolated from clinical specimens associated with various infections, including soft tissue abscesses, septic arthritis, and necrotizing fasciitis [1][2]. It resides within the normal human microbiota, particularly in the gastrointestinal tract and skin, which raises questions regarding its pathogenic potential versus its role as a commensal organism [3][4]. The bacterium expresses multiple surface proteins, some of which may include LPXTG motif proteins that have roles in adhesion and immune evasion [5][6].
The binding capabilities of these proteins, particularly those that can interact with human serum albumin (HSA) [7][8], imply a sophisticated adaptation to the host environment. The PAB (peptostreptococcal albumin-binding) protein identified in F. magna demonstrates a domain with considerable homology to HSA-binding proteins, illustrating its mechanism for immune evasion by sequestering vital host molecules [9]. Moreover, the ability of this bacterium to induce significant inflammatory responses through its secreted proteins underscores its pathogenic mechanisms [3][10].
Furthermore, the presence of LPXTG-motif proteins in F. magna represents an important area of study for understanding the strategies employed by this organism to colonize human tissues effectively and circumvent host defenses, making it a relevant target for research focusing on bacterial virulence factors [5][11]. Continued exploration into the specific functions of the LPXTG-motif anchor proteins in F. magna will enhance our understanding of its pathogenicity and adaptation within the human host.
References:
[1] A. Neumann, L. Björck, & I. Frick. Finegoldia magna, an anaerobic gram-positive bacterium of the normal human microbiota, induces inflammation by activating neutrophils. Frontiers in Microbiology, vol. 11, 2020. https://doi.org/10.3389/fmicb.2020.00065
[2] M. Zou, Z. Yang, et al. Gut microbiota on admission as predictive biomarker for acute necrotizing pancreatitis. Frontiers in Immunology, vol. 13, 2022. https://doi.org/10.3389/fimmu.2022.988326
[3] E. Murphy and I. Frick. Gram-positive anaerobic cocci – commensals and opportunistic pathogens. Fems Microbiology Reviews, vol. 37, no. 4, p. 520-553, 2013. https://doi.org/10.1111/1574-6976.12005
[4] E. Taş and K. Ülgen. Understanding the adhd-gut axis by metabolic network analysis. Metabolites, vol. 13, no. 5, p. 592, 2023. https://doi.org/10.3390/metabo13050592
[5] D. Breidung, S. Delavari, I. Megas, B. Reichert, & M. Billner. Necrotizing fasciitis after panniculectomy caused by finegoldia magna. Plastic and Reconstructive Surgery Global Open, vol. 12, no. 4, p. e5773, 2024. https://doi.org/10.1097/gox.0000000000005773
[6] J. Byeon, K. Blizinsky, et al. Insights into the skin microbiome of sickle cell disease leg ulcers. Wound Repair and Regeneration, vol. 29, no. 5, p. 801-809, 2021. https://doi.org/10.1111/wrr.12924
[7] E. Murphy, M. Mörgelin, D. Reinhardt, A. Olin, L. Björck, & I. Frick. Identification of molecular mechanisms used by finegoldia magna to penetrate and colonize human skin. Molecular Microbiology, vol. 94, no. 2, p. 403-417, 2014. https://doi.org/10.1111/mmi.12773
[8] E. Bormotova and T. Gupalova. The relationship between albumin-binding capacity of recombinant polypeptide and changes in the structure of albumin-binding domain. Bulletin of Experimental Biology and Medicine, vol. 159, no. 3, p. 393-397, 2015. https://doi.org/10.1007/s10517-015-2972-z
[9] P. Samuel, M. Rickard, M. Boob, M. Gruebele, & T. Pogorelov. In silico protein dynamics in the human cytoplasm: partial folding, misfolding, fold switching, and non‐native interactions. Protein Science, vol. 32, no. 11, 2023. https://doi.org/10.1002/pro.4790
[10] S. Lejon, J. Cramer, & P. Nordberg. Structural basis for the binding of naproxen to human serum albumin in the presence of fatty acids and the ga module. Acta Crystallographica Section F Structural Biology and Crystallization Communications, vol. 64, no. 2, p. 64-69, 2008. https://doi.org/10.1107/s174430910706770x
[11] M. Hoeksema, M. Eijk, H. Haagsman, & K. Hartshorn. Histones as mediators of host defense, inflammation and thrombosis. Future Microbiology, vol. 11, no. 3, p. 441-453, 2016. https://doi.org/10.2217/fmb.15.151