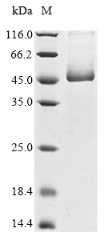
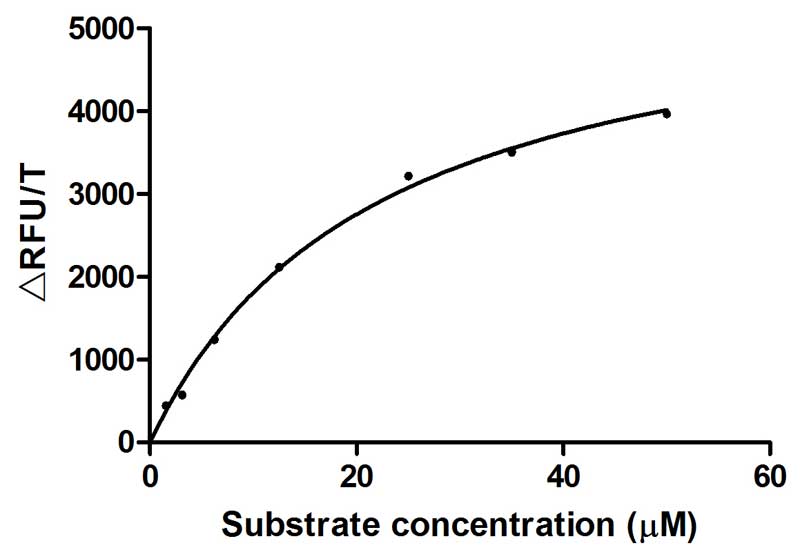
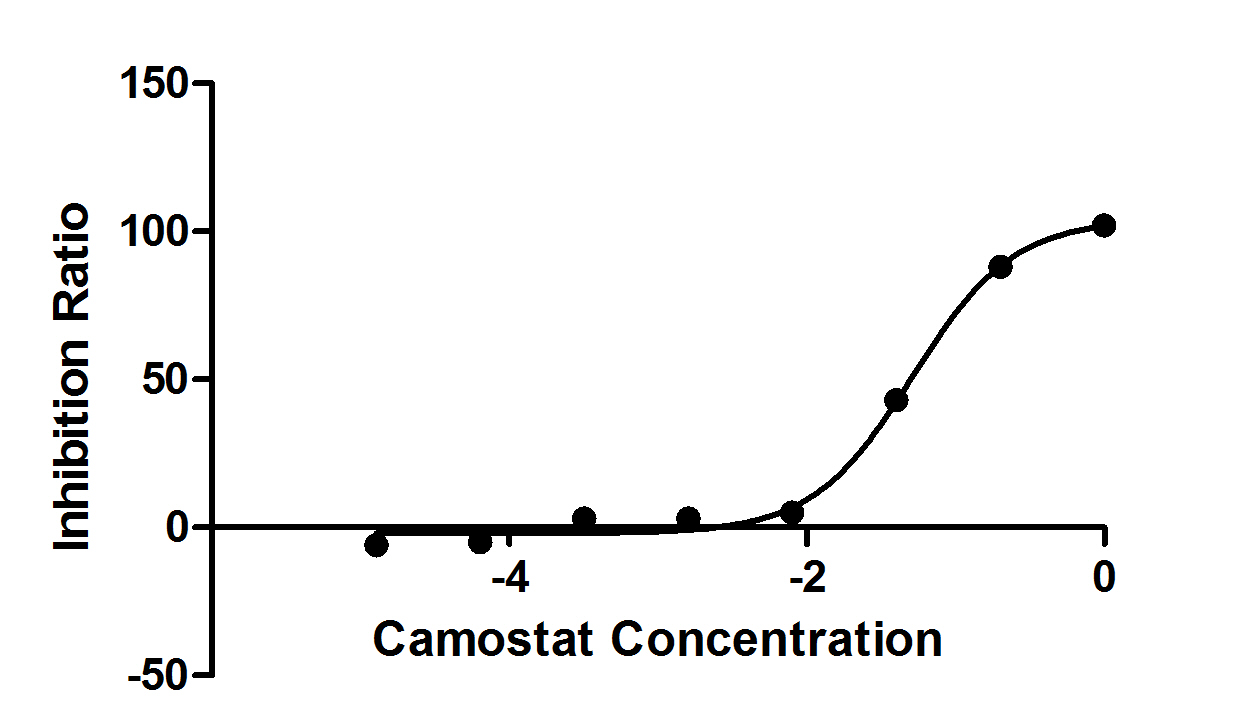
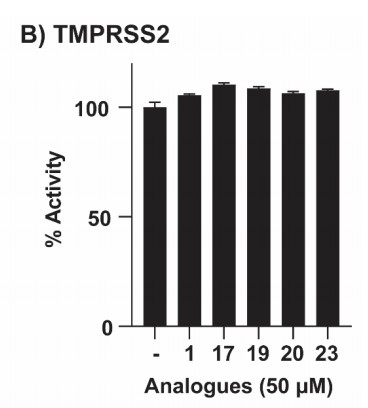
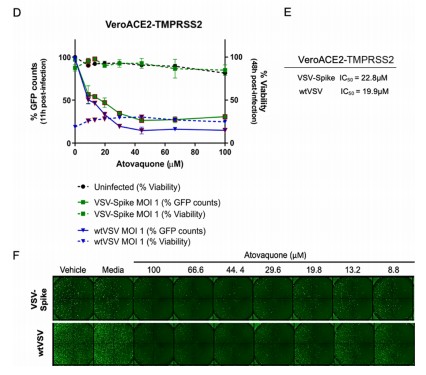
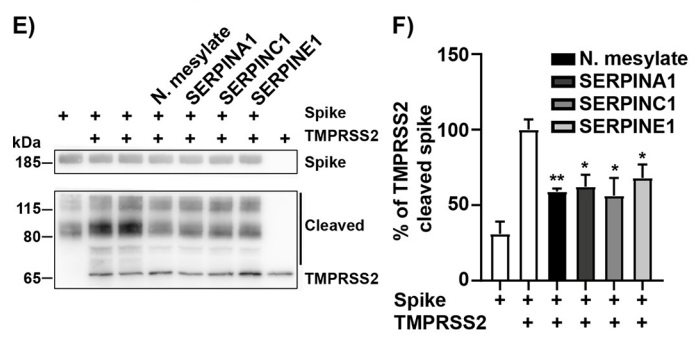
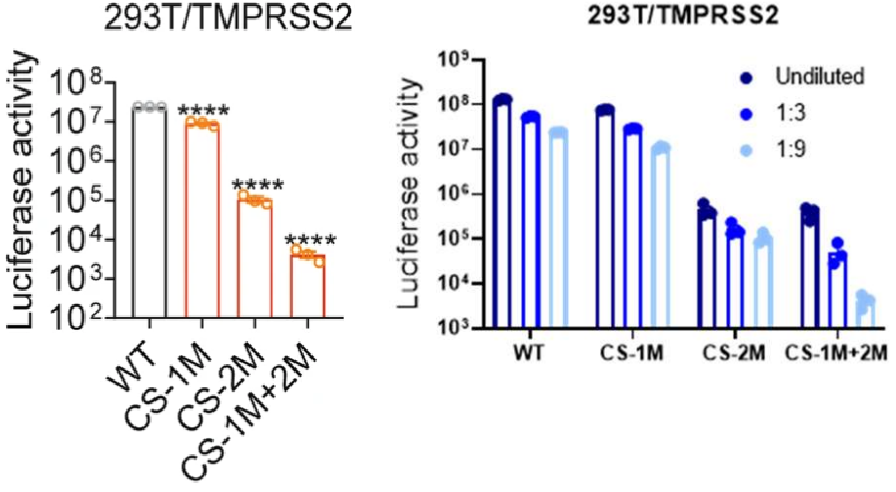
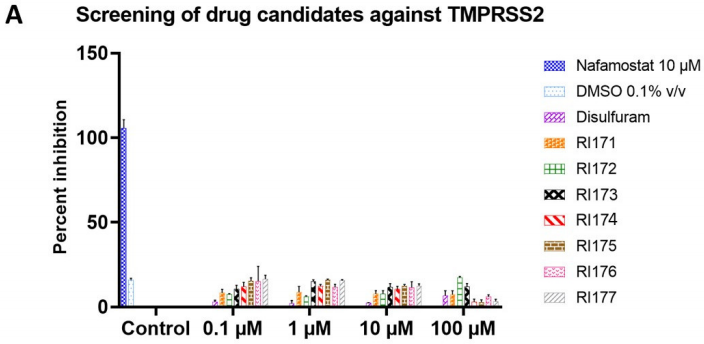
The recombinant active human Transmembrane protease serine 2 (TMPRSS2) is produced by the expression of a target DNA sequence with 6xHis, N-terminal tag(s), in the yeast expression system. The target DNA sequence encodes the 106-492aa region of the human TMPRSS2. The purity of this partial-length protein is greater than 85% determined by SDS-PAGE. The gel showed a molecular weight band of about 45 kDa under reducing conditions. And its enzymatic activity was verified by its ability to cleave fluorogenic peptide substrate (Boc-Gln-Ala-Arg-AMC) (Km is 21.93μM).
The protease TMPRSS2 plays an important role in the infection mechanism of human coronaviruses, such as SARS-CoV and SARS-CoV-2. Cell entry of human coronaviruses depends on the binding of the viral spike (S) glycoprotein to cellular ACE2 receptor and S protein priming by host cell protease TMPRSS2. Moreover, TMPRSS2 protein is commonly used to study cancer. Previous studies have shown that the TMPRSS2 gene was up-regulated by androgenic hormones in prostate cancer cells and down-regulated in androgen-independent prostate cancer tissue.