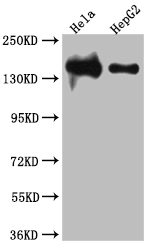
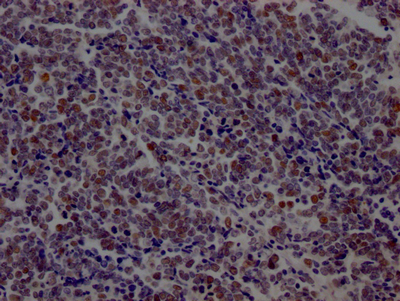
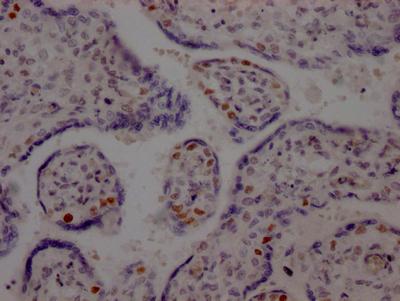
LIG1 is the main replicative ligase involved in DNA replication, recombination, and the base excision repair process. LIG1 is associated with the replisome to finalize Okazaki fragment maturation, completing more than 50 million ligation events during each cycle of DNA replication. It also ligates single-strand DNA breaks during long-patch base excision repair (BER) and can participate in the alternative end-joining (A-EJ) pathway. LIG1 employs a unique metal-mediated fidelity mechanism whereby rigidity imposed by metal binding allows for discrimination against base mismatches on the 3′-hydroxyl side of the nick.
This recombinant LIG1 antibody was developed with the Single B cell platform. The main process included identification and isolation of single B cells; amplification and cloning of LIG1 antibody gene; expression, screening, and identification of antibody specificity. And this LIG1 antibody has been validated in ELISA, WB, IHC.