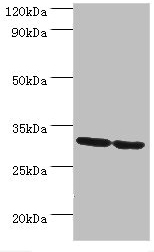
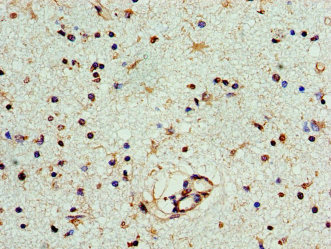
Full Product Name
Rabbit anti-Homo sapiens (Human) PNP Polyclonal antibody
Alternative Names
FLJ94043 antibody; FLJ97288 antibody; FLJ97312 antibody; Inosine phosphorylase antibody; Inosine-guanosine phosphorylase antibody; MGC117396 antibody; MGC125915 antibody; MGC125916 antibody; NP antibody; Np1 antibody; Nucleoside phosphorylase antibody; PNP antibody; Pnp1 antibody; PNPH_HUMAN antibody; PRO1837 antibody; PUNP antibody; Purine nucleoside orthophosphate ribosyltransferase antibody; Purine nucleoside phosphorylase 5a antibody; Purine nucleoside phosphorylase antibody
Immunogen
Recombinant Human Purine nucleoside phosphorylase protein (1-289AA)
Immunogen Species
Homo sapiens (Human)
Purification Method
Antigen Affinity Purified
Concentration
It differs from different batches. Please contact us to confirm it.
Buffer
PBS with 0.02% sodium azide, 50% glycerol, pH7.3.
Tested Applications
ELISA, WB, IHC
Recommended Dilution
| Application |
Recommended Dilution |
| WB |
1:500-1:2000 |
| IHC |
1:20-1:200 |
Storage
Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
Lead Time
Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
Usage
For Research Use Only. Not for use in diagnostic or therapeutic procedures.