In 1984, S Munro and H R Pelham developed a technique to promote the purification and detection of recombinant proteins, in which an oligonucleotide encoding a peptide was fused to the protein of interest and the target protein was detected with a monoclonal antibody that specifically recognizes this peptide [1]. With continuous advancement and evolution of the epitope tagging technique, epitope tagging systems have become a vital tool in many fields of scientific research.
Herein, we will mainly introduce the epitope tag, its applications, the pros and cons of using epitope tag, how to choose an appropriate epitope tag, as well as some frequently used epitope tags.
1. What Is An Epitope Tag?
The epitope tags are 6 to 15-amino acid short peptide sequences that are introduced into the proteins of interest, allowing the purification and detection of the target proteins through their recognition by well-characterized and validated corresponding tag antibodies.
Proteins requiring epitope tagging include those which lack specific antibodies, including newly discovered proteins or poorly immunogenic proteins, and those that have low abundance in endogenous conditions.
Epitope tagging is achieved by fusing the oligonucleotide sequence of the epitope to the encoding sequence of the protein of interest, often on the N- or C-terminus through genetic engineering or chemical conjugation. The epitope tag may also be added to the target protein sequence, such as in the ring structure or in inter-domain regions exposed to solvents.
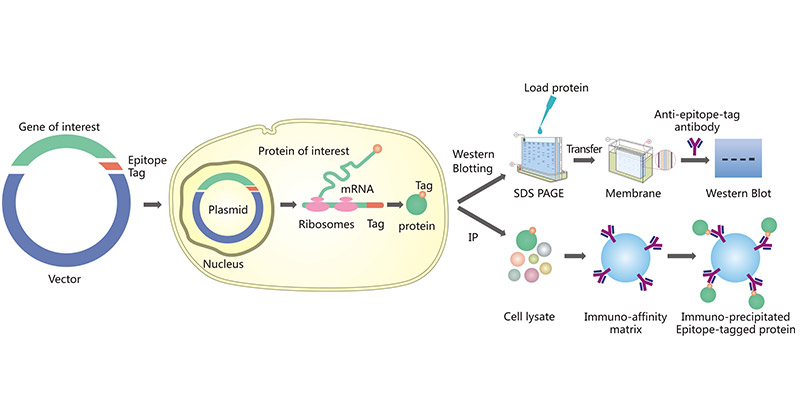
Epitope tagging begins by integrating the epitope tag-encoding gene into the gene coding for the target protein, followed by cloning the fused gene into an expression vector. The constructed expression vector is transformed or transferred into host cell expression systems, including Escherichia coli, yeast, insect, and mammalian cells. The resultant epitope-tagged fusion protein can be either purified or detected via an epitope-tag-specific antibody. The recognition sequence for a protease is frequently added to enable the release of the epitope tag.
Figure 1. Epitope tagging of proteins and common application of epitope tagged proteins
2. What Is An Epitope Tag Used for?
Epitope tags and their corresponding tag antibodies were initially employed for detecting and purifying numerous proteins. Later, epitope tags are used for many biological applications including western blot (WB), immunoprecipitation, immunofluorescence microscopy [2], flow cytometry, protein trafficking in live cells [3][4], protein crystallisation and topology [5].
Epitope-tagged proteins are applied in immunoprecipitation (IP) studies when antibodies against the target protein are unavailable. In co-IP, the epitope-tagged protein is expressed in host cells and gets immunoprecipitation with the antibody specifically against its epitope tag. Proteins that interact with the epitope-tagged protein are co-immunoprecipitated. The resulting protein complex can be analyzed via WB.
Similarily, chromatin immunoprecipitation (ChIP) or RNA immunoprecipitation (RIP) are applied to analyze domains of the genome in which an epitope-tagged DNA-binding protein combines or to identify regions of the genome where an epitope-tagged RNA-binding protein interacts, respectively, using tag-specific antibodies for immunoprecipitation.
During the expression and purification of recombinant proteins, epitope-tagged antibodies are used to improve expression levels, solubility, folding, and purification.
Epitope tags are used in in vitro overexpression studies to investigate and analyze target proteins from multiple aspects, including the monitoring of protein expression, protein localization at the cellular and subcellular levels, and protein purification, as well as protein topology, dynamics and interactions.
In addition, fluorescent reporters, one type of epitope tags, have many applications including visualization of protein localization, acting as transcription reporters (help to analyze transcriptional activity), live cell imaging (track protein dynamics in live cells and thus help to understand cell signaling and to identify proteins participating in various pathways), fluorescence resonance energy transfer (FRET) (investigate interactions between two proteins occurring conformational alterations), fluorescence activated cell sorting (FACS) (separate GFP-expressing cells from cells lacking GFP), and in vivo studies or transgenic tracking (monitor gene expression in various cells and model systems).
3. Advantages and Disadvantages of Using Epitope Tags
Tagging a protein with a preexisting epitope tag is a simple and easy method that enables to identification or purification of proteins promptly. Epitope tagging has many advantages over the use of antibodies generated directly against the target protein [6]. However, this technique also has its own limitations.
|
Advantages
|
Disadvantages
|
|
Enable to the purification or detection of proteins that are difficult to separate and purify via other approaches
|
Should sequence the gene sequence of the target protein to ensure the insertion site of the epitope tag sequence
|
|
Obviating the time and cost related to producing and characterizing antibodies against numerous proteins
|
|
The tag type and insertion site can be chosen in accordance with the needs of the experiment without affecting the target protein's functional regions.
|
The epitope tag may compromise the structure, stability, and function of the target protein
|
|
Epitope tagged-proteins can be easily distinguished and identified from otherwise identical untagged proteins without concern of spurious results resulting from cross-reactive antibodies
|
Heterologous promoters may lead to abnormal expression of the epitope-tagged protein thus causing harmful effects
|
|
Epitope tag-specific antibodies, particularly monoclonal antibodies, can minimize cross-reactivity
|
Epitope tag-bound media (usually monoclonal antibodies are immobilized to chromatographic chromatography resins) are expensive for purification and are not suitable for large-scale protein preparation compared to other affinity purification media
|
|
Epitope tagging enables the detection of in vitro-mutagenized variants in the setting of natural wild-type proteins or the discrimination of individual members of intimately related protein families
|
|
One or more epitope tag can be added to the target protein, increasing the detection limit or assay sensitivity
|
Eluting the target protein with the short peptide corresponding to the epitope tag, low
PH, or other methods such as salts, polyols, and the chelation of tag-bound calcium ions. Some of these methods may appear drastic compared to other affinity purification methods
|
|
Can be applied to study protein-protein interactions, functions, and topology using epitope tag-specific antibodies
|
4. How to Choose An Epitope Tag?
One epitope tag cannot be optimal with respect to all of the parameters of a protein including the yield, solubility, and the folding of its fusion partners. Each epitope tag has its own advantages and disadvantages, so the choice of the most suitable epitope tag is dependent on the experiments. Besides, selecting an epitope tag also needs to take into account many other factors, including the tag's size and charge, downstream analysis, the solubility of the protein, and whether the tag may affect protein function.
Among these factors, the downstream application of the target protein is the most important factor when choosing an epitope tag.
For instance, epitope tags like 6*His, GST (Glutathione-S-transferase), V5, and MBP (Maltose-Binding Protein) are widely used if the desired downstream application is recombinant protein purification.
Fluorescent reporters including Green Fluorescent Protein (GFP), Allophycocyanin (APC), and Fluorescein Isothiocyanate (FITC) are applied for cellular tracking and localization analyses.
FLAG, c-myc, and Hemagglutinin (HA) are frequently used in immunoprecipitation experiments to monitor and study interactions between different proteins or proteins and DNA or RNA.
Sometimes, combinatorial epitope tagging may be used in certain conditions to optimize the downstream applications of the target protein.
5. List of Epitope Tags
There are numerous available epitope tags. They have own strength and drawbacks. Here are list some frequently used epitope tags.
|
Epitope tags
|
Sequence/Size
|
Source
|
Application
|
Pros and cons
|
|
FLAG
|
DYKDDDDK
|
Synthetic
|
Protein detection, protein-protein interactions through IP analysis
|
Contains internal Enterokinase cleavage site; demonstrated to undergo a post-translational modification that disrupts the anti-tag antibody/tag interactions decreasing the purification yield [7]
|
|
6*His
|
HHHHHH
|
Synthetic
|
Affinity purification, protein detection
|
The most commonly used purification tag; Purification via Nickel, Cobalt, Copper, or Zinc column
|
|
HA
|
YPYDVPDYA
|
Amino acids 98-106 of Human influenza hemagglutin
|
Protein detection, protein-protein interactions through IP analysis
|
Strong immunoreactive epitope and mild elution conditions; not suitable for detection or purification of apoptotic cell-deriving proteins because HA can be cleaved by CASP3 and CASP7 after its sequence DVPD, resulting in immunoreactivity lose
|
|
c-myc
|
EQKLISEEDL
|
Amino acid residues 410-419 of Human c-Myc
|
Protein detection, protein-protein interactions through IP analysis
|
Successfully applied in WB hybridization technology, immunoprecipitation (IP) and flow cytometry; low pH elution condition of myc recombinant protein tends to reduce protein viability
|
|
GST
|
27 kDa
|
Eukaryotes and Prokaryotes
|
Affinity purification, protein detection, protein-protein interactions
|
Affinity purification via relatively reusable glutathione conjugated columns; purification under native conditions only; highly antigenic; may cause insolubility and dimerize
|
|
Strep-tag II
|
WSHPQFEK
|
Synthetic
|
Protein detection
|
Exhibits intrinsic affinity toward streptavidin and can be fused to recombinant proteins in various fashions; its neutral pI has little effect on protein function or folding; Strep-tag II and Strep-Tactin resin highly specific interaction enhances the purity of the recombinant protein ; suitable for the production of biologically active proteins because it allows the elution of recombinant protein under gentle, physiological conditions [8]
|
|
MBP
|
43 kDa
|
Maltose/maltodextrin system of E. coli
|
Affinity purification, protein detection, protein-protein interactions
|
Can stabilize, solubilize and even crystallize recombinant proteins that are fused to it; greatly enhances proteins stability, solubility, and production [9]
|
|
S-tag
|
S-peptide
KETAAAKFERQHMDS
|
15 amino acid sequence at N-terminus of RNase A
|
Protein detection
|
Not recommended for affinity purification as protein properties may change during low pH elution step
|
|
E-Tag
|
GAPVPYPDPLEPR
|
Bone hormone osteocalcin produced by osteoblasts
|
Protein detection, protein-protein interactions in IP analysis
|
|
|
Avi-tag
|
CGLNDIFEAQKIEWHE
|
Synthetic
|
Protein detection
|
Allows for enzymatic biotinylation by the biotin ligase BirA from E. coli; May decrease solubility
|
|
Calmodulin-binding protein (CBP)
|
CBP peptide/
KRRWKKNFIAVSAANRFKKISSSGAL
|
Cytoplasm of all eukaryotic cells
|
Protein detection
|
No cross-reaction with any endogenous E.coli proteins; small size of CBP (4 kDa) makes it ideal for purifying delicate proteins under mild conditions; has the relatively high affinity for calmodulin (CaM); Not useful for purification from eukaryotic cells; N-terminus tagging may reduce translation efficiency
|
|
Green fluorescent protein (GFP)
|
27 kDa
|
Jellyfish Aequorea Victoria
|
Protein detection ands visualization, fluorescence resonance energy transfer (FRET), live cell imaging, fluorescence activated cell sorting (FACS)
|
Major excitation peak at a wavelength of 395 nm and a minor one at 475 nm; emission peak is at 509 nm; Detectable in living organisms without antibody; Can sometimes be non-specifically directed to nucleus; Useful to monitor protein-protein interactions by FRET; Very large; Dimerization may occur; Useful in protein-folding assays
|
|
Red Fluorescent Protein (RFP)
|
26 kDa
|
Isolated from Discosoma
|
Protein detection ands visualization, fluorescence resonance energy transfer (FRET), live cell imaging, fluorescence activated cell sorting (FACS)
|
The excitation maximum is 558 nm, and the emission maximum is 583 nm; Unusable for many experiments that take place in a shorter time frame because its maturation time is about 24 hours; its tetrameric form can affect the function of the target protein
|
|
mCherry
|
28 kDa
|
Disc corals of Discosoma species
|
Protein detection ands visualization, fluorescence resonance energy transfer (FRET), live cell imaging, fluorescence activated cell sorting (FACS)
|
Useful to monitor protein-protein interactions by FRET; Excitation maximum at 587 nm and an emission maximum at 610 nm; resistant to photobleachin; quite stable and have an extremely rapid maturation rate
|
|
HSV
|
SQPELAPEDPED
|
Herpes Simplex Virus tag
|
Protein detection
|
C-terminal placement only; Not recommended for affinity purification as protein properties may change during low pH elution step
|
|
Fc-Fusion Proteins
|
|
Fc domain of IgG Antibody
|
Protein detection, affinity purification
|
Can increase the solubility and stability of the tagged protein both in vitro and in vivo; Easy cost-effective purification by protein-G/A affinity chromatography [10]
|
|
Beta-Galactosidase
|
116 kDa
|
Enzyme coded by lac z gene in the lac operon of E. coli
|
Protein detection
|
Confers the enzyme activity to the bound protein and thus allows for enzymatic protein quantification assays; May protect from proteolytic activity; May decrease solubility and form tetramers in solution
|
|
VSV-G
|
YTDIEMNRLGK
|
11 amino acid C-terminus of the Vesicular Stomatitis viral glycoprotein
|
Protein detection
|
Not recommended for affinity purification as protein properties may change during low pH elution step
|
|
Thioredoxin
|
12 kDa
|
Found in all organisms; Encoded by TXN and TXN2 genes in humans
|
Protein detection
|
Can increase the solubility of tagged protein
|
|
AU1
|
DTYRYI
|
Major capsid protein of bovine papillomavirus-1 (BPV-1)
|
Protein detection
|
Not recommended for affinity purification as protein properties may change during low pH elution step
|
|
AU5
|
TDFYLK
|
Major capsid protein of bovine paillomavirus-1 (BPV-1)
|
Protein detection
|
Not recommended for affinity purification as protein properties may change during low pH elution step
|
|
V5
|
GKPIPNPLLGLDST
|
Amino acid residues 95 to 108 of RNA polymerase alpha subunit of simion virus 5
|
Protein detection, protein affinity purification, protein-protein interactions through IP analysis
|
Some cross reactivity may occur when using mammalian expression systems; Recommended for affinity purification in combination with His-tag
|
|
T7
|
MASMTGGQQMG
|
11 amino acid N-terminus of Bacteriophage T7 gene 10
|
Protein detection
|
May increase protein expression; Not recommended for affinity purification as protein properties may get altered during low pH elution step
|
|
KT3
|
KPPTPPPEPET
|
Simian Virus 40 (SV40) large T-antigen
|
Protein detection
|
Not recommended for affinity purification as protein properties may get altered during low pH elution step
|
|
TK15
|
(R/K)TV(I/L)HGESNSA(I/L)(I/L)(I/L)GPR
|
Xenopus Orc1p
|
Protein detection
|
No cross-reactivity with mammalian or bacterial proteins; Used for immunoaffinity purification
|
CUSABIO offers some premium monoclonal antibodies against commonly used epitope tags. These epitopes tag antibodies undergo protein A/G affinity purification and have high specificity.
|
Epitope-tagged Antibodies
|
Applications
|
|
6*His Monoclonal Antibody
|
ELISA, WB
|
|
GFP Monoclonal Antibody
|
ELISA, WB, IP
|
|
Myc tag Monoclonal Antibody
|
ELISA, WB, IF, IP
|
|
GST Monoclonal Antibody
|
ELISA, WB
|
|
E-Tag Monoclonal Antibody
|
ELISA, WB, IP
|
|
HA-Tag Monoclonal Antibody
|
ELISA, WB, IF, IP, FC
|
|
Flag Tag Monoclonal Antibody
|
ELISA, WB, IF, IP
|
|
MBP Monoclonal Antibody
|
ELISA, WB
|
|
Avi-Tag Monoclonal Antibody
|
ELISA, WB
|
|
CBP Tag Monoclonal Antibody
|
ELISA, WB
|
|
HSV-Tag Monoclonal Antibody
|
ELISA, WB
|
|
V5-Tag Monoclonal Antibody
|
ELISA, WB
|
|
S-Tag Monoclonal Antibody
|
ELISA, WB
|
|
Strep-Tag Monoclonal Antibody
|
ELISA, WB
|
|
KT3-Tag Monoclonal Antibody
|
ELISA, WB
|
|
T7-Tag Monoclonal Antibody
|
ELISA, WB
|
|
mCherry-Tag Monoclonal Antibody
|
ELISA, WB
|
|
VSV-G-Tag Monoclonal Antibody
|
ELISA, WB, IF, IP
|
|
RFP-Tag Monoclonal Antibody
|
ELISA, WB
|
References
[1] Munro S & Pelham HRB (1984). Use of peptide tagging to detect proteins expressed from cloned genes: deletion mapping functional domains of Drosophila hsp 70 [J]. EMBO J 3, 3087–3093.
[2] Götzke H, Kilisch M, et al. (2019). The ALFA-tag is a highly versatile tool for nanobody-based bioscience applications [J]. Nat Commun 10, 4403.
[3] Kocaoglu O & Carlson EE (2016). Progress and prospects for small-molecule probes of bacterial imaging [J]. Nat Chem Biol 12, 472–478.
[4] Nooh MM & Bahouth SW (2017). Visualization and quantification of GPCR trafficking in mammalian cells by confocal microscopy [J]. In Methods in Cell Biology (Shukla AK, ed), pp. 67–78. Academic Press, Cambridge, MA.
[5] Tamura R, Oi R, Akashi S, Kaneko MK, Kato Y & Nogi T (2019). Application of the NZ-1 Fab as a crystallization chaperone for PA tag-inserted target proteins [J]. Protein Sci 28, 823–836.
[6] Christian E. Fritze and Thomas R. Anderson. Epitope tagging: General method for tracking recombinant proteins [J]. Methods in Enzymology Volume 327, 2000, Pages 3-16.
[7] Schmidt PM, Sparrow LG, et al. (2012) Taking down the FLAG! How insect cell expression challenges an established tag-system [J]. PLoS One 7, e37779.
[8] Sumreet Singh Johar and Joey N. Talbert. Strep-tag II fusion technology for the modification and immobilization of lipase B from Candida antarctica (CALB) [J]. J Genet Eng Biotechnol. 2017 Dec; 15(2): 359–367.
[9] Momin, A.A., Hameed, U.F.S. & Arold, S.T. Passenger sequences can promote interlaced dimers in a common variant of the maltose-binding protein [J]. Sci Rep 9, 20396 (2019).
[10] Daniel M Czajkowsky, Jun Hu, et al. Fc-fusion proteins: new developments and future perspectives [J]. EMBO Mol Med. 2012 Oct; 4(10): 1015–1028.
CUSABIO team. Epitope Tags Overview. https://www.cusabio.com/c-21099.html



Comments
Leave a Comment