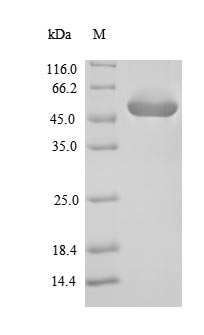
Recombinant Enterococcus faecalis Gelatinase (gelE) gets expressed in E.coli and comes with an N-terminal 6xHis-SUMO tag that makes purification straightforward. The product appears to represent the full length of the mature protein, spanning amino acids 193-510. SDS-PAGE analysis confirms it reaches purity levels greater than 90%, which likely ensures minimal contamination for precise experimental outcomes. This product is strictly for research use only and should not be used in diagnostic or therapeutic applications.
Gelatinase from Enterococcus faecalis is a notable metalloprotease that breaks down gelatin and other proteins. Research suggests it's involved in bacterial virulence and biofilm formation. This makes it an important target for studying bacterial pathogenicity and antibiotic resistance mechanisms. The enzyme's ability to degrade extracellular matrix components may highlight its value in infectious disease research and microbial ecology studies.
Potential Applications
Note: The applications listed below are based on what we know about this protein's biological functions, published research, and experience from experts in the field. However, we haven't fully tested all of these applications ourselves yet. We'd recommend running some preliminary tests first to make sure they work for your specific research goals.
Based on the provided information, the folding state and bioactivity of this recombinant GelE protein are unknown and cannot be assumed. Gelatinase is a zinc-metalloprotease that requires precise folding around an active site and correct coordination of metal ions for catalytic activity. While the protein represents the mature enzyme sequence (193-510aa), its expression in E. coli with a large N-terminal 6xHis-SUMO tag creates significant uncertainty about its native conformation. The SUMO tag may enhance solubility but does not guarantee correct folding of the protease domain. Therefore, any application that depends on the protein's specific enzymatic activity or its properly formed substrate-binding site is speculative without experimental validation.
1. Protein-Protein Interaction Studies Using Pull-Down Assays
The N-terminal 6xHis-SUMO tag allows this recombinant GelE to be immobilized for pull-down assays. However, the utility of these assays for identifying biological binding partners is entirely contingent on the protein being correctly folded. If the protein is misfolded, it may not present the correct binding interfaces, leading to the identification of non-specific interactors. Such experiments could be conducted, but results must be interpreted as preliminary and require confirmation with a validated, active protein.
2. Antibody Development and Immunoassay Applications
This recombinant GelE protein is suitable for use as an immunogen to generate specific antibodies. The mature protein sequence increases the likelihood that antibodies will recognize epitopes present in the native enzyme. However, it is important to note that antibodies generated will primarily recognize linear epitopes. Their ability to bind the natively folded, active gelatinase in Enterococcus faecalis cultures is not guaranteed and requires empirical validation (e.g., using culture supernatants from a gelE+ strain). The protein is excellent for developing ELISA assays to detect anti-GelE antibodies.
3. Biochemical Characterization and Substrate Specificity Studies
This purified recombinant GelE protein is a starting point for biochemical characterization. Studies on substrate binding site analysis and binding kinetics with potential substrates cannot be meaningfully performed without first confirming the protein's enzymatic activity. It should first perform a gelatin zymography or a fluorescence-based protease activity assay to confirm functionality. If active, then studies on kinetics, pH optimum, and substrate specificity can be pursued. The initial studies should focus on biophysical properties (e.g., thermal stability via differential scanning fluorimetry) that are independent of activity.
4. Comparative Proteomics and Evolutionary Studies
The recombinant GelE protein can serve as a reference standard for comparative studies. This application is valid for specific, non-activity-dependent comparisons. It is suitable as a standard for mass spectrometry-based identification or for comparing immunological reactivity across strains. However, it cannot be used for "comparing expression levels" of the active enzyme in different isolates, as the assay would detect immunoreactivity, not necessarily functional enzyme levels.
Final Recommendation & Action Plan
The immediate and essential first step is to experimentally validate the proteolytic activity of this recombinant GelE protein using a functional assay, such as gelatin zymography or a fluorescence-quenched substrate assay. The outcome of this test is critical and dictates all subsequent applications: a positive result would justify and enable functional studies in Applications 1 (interactions) and 3 (kinetics/substrate specificity). A negative result would limit its reliable use to Applications 2 (antibody production, with the caveat about native conformation recognition) and 4 (as a reference antigen for comparative immunology or MS). Therefore, the activity assay is a prerequisite before investing resources in binding or kinetic studies, as data generated without this confirmation would be uninterpretable. The SUMO tag can be cleaved off to assess its impact on activity and folding.