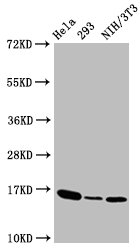
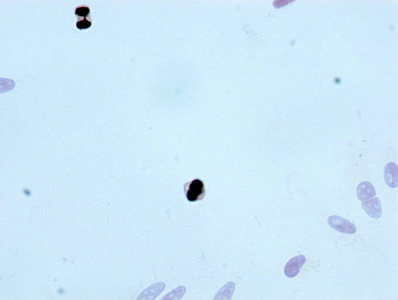
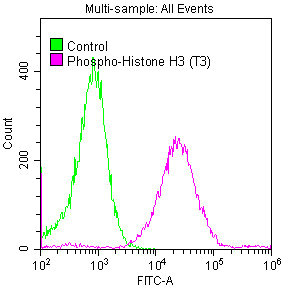
The process of producing the phospho-Histone H3.3 (T3) recombinant antibody commences with the cloning of the genes encoding the H3F3A antibody, encompassing both heavy and light chains, and their insertion into expression vectors. These modified vectors are then introduced into host cells through transfection, prompting the host cells to take on the role of antibody production and secretion. The resulting phospho-Histone H3.3 (T3) antibody is purified using affinity chromatography to ensure its purity and effectiveness. Rigorous testing follows to evaluate its functionality across a spectrum of applications, including ELISA, WB, ICC, and FC, all designed for the specific detection of the human and mouse H3F3A proteins phosphorylated at T3.
Phosphorylation of Histone H3.3 at threonine 3 (T3) is involved in transcriptional regulation, chromatin remodeling, DNA repair, cell cycle regulation, epigenetic signaling, and cellular memory, and has implications in various diseases. It is a dynamic modification that helps regulate gene expression and chromatin structure.