In the eukaryotic nucleus, histone H2A and H2B interact closely to form dimers, H3 and H4 form tetramers, and two H2A, H2B dimers and one H3 and H4 tetramer form core histone octamers. DNA entangles with the core histone octamer to form a nucleosome, which is the basic unit of chromosomes. Histone is a powerful artifact of DNA compression. Post-translational modification of histones is a common pathway for the regulation of nucleosomal structures. Four frequently common histone modifications include methylation, acetylation, phosphorylation, and ubiquitination.
Methylation of histones is closely related to gene transcriptional activation, transcriptional silencing, X chromosome inactivation, and the heterochromatin dense state. Histone methylation was thought to be an irreversible process for a long period. With the discovery of the first histone demethylase in 2004 [1], it is clear that histone methylation is a dynamically regulated process. Histone methylation is regulated by histone methyltransferases (HMTs) and histone demethylases (HDMs). Histone demethylation is the opposing process to the methylation process.
1. What Is Histone Demethylation?
Histone demethylation is a process during which histone demethylases (HDMs) catalyze the removal of methyl groups from specific amino acids in the N-terminal tails of histones. Histone demethylases mainly act on lysine (K) residues on histone H3, including K4, K9, K27, and K36. Histone demethylases are also called histone lysine demethylases (KDMs).
2. Two Histone Demethylase Families: LSD and JmjC
Two evolutionarily conserved histone demethylase families have been identified: lysine-specific demethylase (LSD) and Jumonji C (JmjC) domain-containing demethylase (JHDM) [1][2]. They use different mechanisms to remove methyl groups.
2.1 LSD Demethylases
LSD family consists of LSD1 and LSD2, which demethylate monomethylated and dimethylated lysine residues through flavin adenine dinucleotide (FAD)-dependent amine oxidase reaction. LSD1, also known as KDM1A or AOF2, is the first discovered histone lysine demethylase that removes monomethyl and dimethyl groups from lysine 4 in histone H3 (H3K4me1/2) or lysine 9 in histone H3 (H3K9me1/2), acting as a repressor or activator of gene expression, respectively.
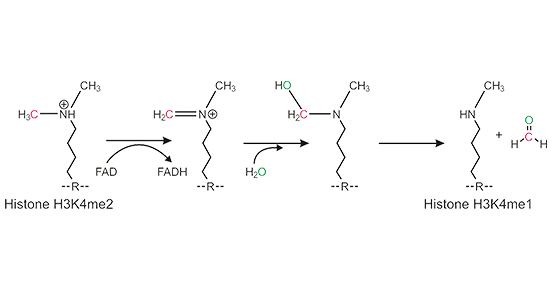
LSD1 Demethylation Mechanism:
LSD1 contains a flavin adenine dinucleotide (FAD)-dependent amine oxidase domain that is responsible for its demethylase activity. LSD1 uses FAD as a co-factor to oxidize the methenyl group bound to the substrate through an amine oxidation reaction, generating an iminium (N+) ion. The iminium (N+) ion is spontaneously hydrolyzed and generates chlorophenopyranolamine, which is unstable and degrades to release a formaldehyde molecule, leading to a mono-methylated lysine (H3K4me1). LSD1 can not demethylate trimethylated lysine residues because it requires a protonated nitrogen as a hydrogen donor, and the nitrogen of a trimethyl-lysine residue is not protonated [3].
Figure 1: LSD1 Demethylation Mechanism
This picture is cited from: https://journals.biologists.com/dev/article/136/6/879/43849/Developmental-roles-of-the-histone-lysine
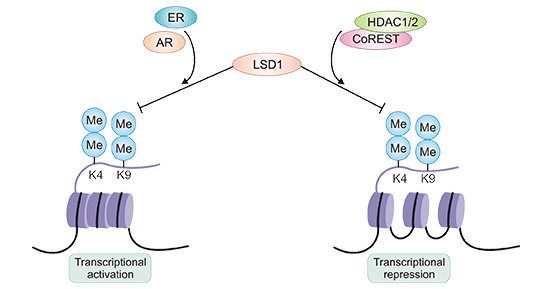
LSD1 interacts with CoREST and histone deacetylase 1/2 (HDAC1/2), which confers more specificity for H3K4me1/2, leading to its demethylation thus promoting heterochromatin formation and transcriptional inactivation of the target genes [1][4]. However, LSD1 acting as a transcriptional activator demethylates H3K9me1/2 when it is complexed with activated androgen receptors such as estrogen receptors (ERs) or androgen receptors (ARs) [5][6].
Figure 2: LSD1 acts as both transcriptional activator or repressor
This picture is sourced from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5691370/
By demethylating H3K4, LSD1 makes DNMT3L, a positive regulator of DNA methyltransferase (DNMTs), bind to the unmethylated K4 site, which promotes the expression of DNMTs, causing DNA re-methylation thus leading to gene transcription repression.
In addition, LSD1 can also demethylate non-histone proteins, such as K370me1 and K370me2 on p53, Lys1096 on DNMT1, as well as Lys185 on E2F1.
LSD2, also referred to as KDM1B or AOF2, is another FAD-dependent amine oxidase homolog containing a SWIRM domain and specifically targets H3K4me1/2 [7]. KDM1B, however, is unable to form a complex with CoREST as it lacks a tower domain.
2.2 JmjC Demethylases
Tsukada et al. identified the first JmjC-domain-containing protein JHDM1A/KDM2A as an H3K36me1/2 demethylase in 2006 [2]. According to the sequence information, proteins containing JmjC can be divided into 7 families, namely JHDM1, JHDM2, JHDM3, JARID1, UTX/UTY, PHF8, and proteins containing only the JmjC domain. The JmjC family consists of 30 members, nearly 20 of which have so far been shown to have histone demethylase activity.
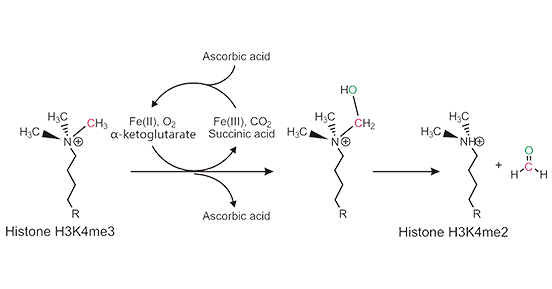
JmjC Demethylation Mechanism:
The JmjC demethylases belong to the dioxygenase superfamily and use a demethylation mechanism different from that of LSD1. JmjC family-mediated demethylation is a ferrous ion and α-ketoglutarate-dependent hydroxylase reaction (2OG-Fe(II)-dependent dioxygenase reaction). The ferrous ion and α-ketoglutarate are required to act coordinately to hydroxylate the methyl motif, creating an unstable carbinolamine group that is spontaneously released as formaldehyde. JmjC demethylases can remove mono-methylated, di-methylated, and trimethylated lysine residues.
Figure 3: JmjC Demethylation Mechanism
This picture is sourced from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5691370/
Table 1. HDM family Members
| Family |
HDM Name |
Structural Domain |
Histone Substrate |
Function |
| LSD |
LSD1 |
KDM1A |
SWIRM, AOL, Tower domains |
H3K4me1/2, H3K9me1/2 |
Transcription repression or activation; DNA methylation; early embryonic development |
| LSD2 |
KDM1B |
AOL and SWIRM domains |
H3K4me1/2 |
Associated with transcribed coding regions in genes and is important for optimal gene transcription |
| JmjC domain-containing protein |
JHDM1 |
JHDM1A/KDM2A |
JmjC, LRRS, F-box, CXXX zinc finger, and DNA-binding domains |
H3K36me1/2 |
Transcription elongation |
| JHDM1AB/KDM2B |
H3K36me1/2; H3K4me3 |
Transcription elongation; anti-oncogene |
| JHDM2 |
JHDM2A/KDM3A |
JmjC, Zinc-like finger domain |
H3K9me1/2 |
Androgen receptor activation; spermatogenesis; diabetes |
| JHDM2B/KDM3B |
H3K9me1/2 |
Anti-oncogene |
| JHDM3 |
JHDM3A/JMJD2A/KDM4A |
JmjN domain, the JmjC domain, the C-terminal domain, and a zinc-finger motif |
H3K9me2/3; H3K36me2/3 |
Transcription repression; genome integrity maintenance |
| JHDM3B/JMJD2B/KDM4B |
H3K9me2/3; H3K36me 2/3 |
Heterochromatin formation |
| JHDM3C/JMJD2C/KDM4C |
H3K9me2/3; H3K36me2/3 |
Oncogene; maintenance of totipotency of stem cells |
| JHDM3D/JMJD2D/KDM4D |
H3K9me2/3; H3K36me2/3 |
Androgen receptor activation |
| JARID1 |
JARID1A/KDM5A |
JmjC, JmjN, AT rich interactive, C5HC2 zinc finger, and PHD domains |
H3K4me2/3 |
RB-binding protein |
| JARID1B/KDM5B |
H3K4me1/2/3 |
Transcription repression; oncogene |
| JARID1C/KDM5C |
H3K4me2/3 |
X chromosome- linked intellectual disability |
| JARID1D/KDM5D |
H3K4me2/3 |
Male specific antigen |
| KDM6A |
UTX |
JmjC, six tetratricopeptide repeat (TPR) domains |
H3K27me2/3 |
Regulation of HOX gene expression; anti-oncogene |
| KDM6B |
JMJD3 |
JmjC domain, a C-terminal segment embedded with a GATAL domain |
H3K27me2/3 |
Nerve and epidermal cells develop immunity |
| JMJD6 |
JmjC domain, three apparent nuclear localization signals (NLS), a DNA binding domain (AT-hook domain), a putative sumoylation site, and a polyserine (polyS) domain |
H3K2me2, H4K3me2 |
Transcriptional activation; embryonic development |
3. What Is the Function of Histone Demethylases?
Since the first histone demethylase was discovered in 2004, researchers have fully characterized the activities of the identified histone demethylases and found that these histone demethylases not only can target both histones and non-histone substrates but also play important roles in cancer, development, metabolic diseases (diabetes), and other processes.
3.1 Histone Demethylases and Cancer
LSD1 is abnormally expressed in many cancers and hinders cancer cell differentiation, promotes cancer cell proliferation, metastasis & invasion, and is associated with poor prognosis. Overexpression of LSD1 has been found in hematopoietic and lymphatic neoplasms, including acute myeloid leukemia (AML), acute lymphoblastic leukemia, T-cell non-Hodgkin lymphoma, and Hodgkin lymphoma. LSD1 is also linked to several types of solid tumors, including non-small cell lung carcinoma, neuroblastoma, pancreatic, prostate, and breast cancers. Inhibition of the effects of LSD1 might reduce or block cell growth in these tumors.
Wan, W. et al. found that JMJD1A association with HIF-1α affected the H3K9me2 level in the HRE region of PGK1 promoter in the glycolytic pathway of bladder tumor cells, thereby affecting the expression of PGK1 and further regulating the glucose metabolism pathway of bladder tumor cells, ultimately playing a regulatory role in the growth and proliferation of bladder tumor cells [8].
3.2 Histone Demethylases and Development
A flurry of studies has demonstrated the specific biological roles of histone demethylases in development across species, including in germline maintenance and meiosis, early embryonic development and differentiation, and hormone receptor-mediated transcriptional modulation.
In metazoans, LSD1 acts on H3K4me1/2 and plays important roles in the germline. Several demethylases participate in the progression of pluripotent progenitor cell types into differentiated cell lineages during development. At the blastula stage, LSD1 is both maternally stored and expressed during embryogenesis [9]. Bisguanidine 1c, the LSD1 demethylase inhibitor was used in vitro fertilized mouse embryos, leading to the global H3K4me2 increase and the induction of irreversible arrest in mouse embryos [10]. This demonstrated that LSD1 has an embryonic role and is essential for early differentiation events [10]. In flies, loss of LSD1 results in embryonic lethality.
Both JMJD2C and JHDM2A have been recently implicated in the maintenance of pluripotency in mouse embryonic stem (ES) cells. These two demethylases are expressed in undifferentiated ES cells and appear to remain pluripotency by activating the transcription of pluripotency-maintaining proteins, such as OCT4, NANOG, and TCL1 [11][12].
Jepsen et al. found that JMJD3 acts as a transcriptional target of the silencing mediator for retinoid and thyroid hormone receptors (SMRT) when mouse embryonic neural stem cells (NSCs) were exposed to retinoic acid (RA) to induce differentiation [13]. They also revealed that JMJD3 overexpression in an NSC culture stimulated the expression of neuronal subtype genes such as Dlx5, providing evidence for JMJD3's role in driving neuronal differentiation.
Yamane et al. showed that both LSD1 and JHDM2A co-regulated the androgen receptor (AR)-responsive genes encoding prostate-specific antigen (PSA) and NKX3.1 [6][14].
3.3 Histone Demethylases and Diabetes
Tateishi et al. found that JHDM2A knockout mice showed symptoms of obesity and hyperlipidemia. Detailed studies have shown that the stimulation of β-adrenaline can induce the expression of the JHDM2A gene, which can bind to Ppara and Ucp1 genes to directly regulate their expression. Therefore, the deficiency of the JHDM2A gene affects glycerol release and oxygen metabolism in brown adipose tissue and decreases adipose oxidation and glycerol release in bone tyromuscle, which ultimately leads to obesity and hypertension.
CUSABIO provides various recombinant histone demethylases used for research:
References
[1] Shi Y, Lan F, et al. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1 [J]. Cell 119: 941–953.
[2] Tsukada Y, Zhang Y, et al. (2006) Histone demethylation by a family of JmjC domain-containing proteins [J]. Nature 439: 811–816.
[3] Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy [J]. Nat Rev Drug Discov. 2013;12:917–930.
[4] Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation [J]. Nature. 2005;437:432–5.
[5] Wissmann M. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression [J]. Nat Cell Biol. 2007;9:347–353.
[6] Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription [J]. Nature. 2005;437(7057):436–9.
[7] Ciccone DN. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints [J]. Nature. 2009;461:415–418.
[8] Wan, W., Peng, K., et al. Histone demethylase JMJD1A promotes urinary bladder cancer progression by enhancing glycolysis through coactivation of hypoxia inducible factor 1α [J]. Oncogene 36, 3868–3877 (2017).
[9] McGraw, S., Vigneault, C. and Sirard, M. A. (2007). Temporal expression of factors involved in chromatin remodeling and in gene regulation during early bovine in vitro embryo development [J]. Reproduction 133, 597-608.
[10] Shao, G. B., Ding, H. M. and Gong, A. H. (2008). Role of histone methylation in zygotic genome activation in the preimplantation mouse embryo [J]. In Vitro Cell Dev. Biol. Anim. 44, 115-120.
[11] Katoh, Y. and Katoh, M. (2007). Comparative integromics on JMJD2A, JMJD2B and JMJD2C: preferential expression of JMJD2C in undifferentiated ES cells [J]. Int. J. Mol. Med. 20, 269-273.
[12] Loh, Y. H., Zhang, W., et al. (2007). Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 21, 2545-2557.
[13] Jepsen, K., Solum, et al. (2007). SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron [J]. Nature 450, 415-419.
[14] Yamane, K., Toumazou, C., et al. (2006). JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor [J]. Cell 125, 483-495.
CUSABIO team. Histone demethylases: LSD and JmjC Families. https://www.cusabio.com/c-20140.html




Comments
Leave a Comment