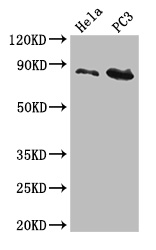
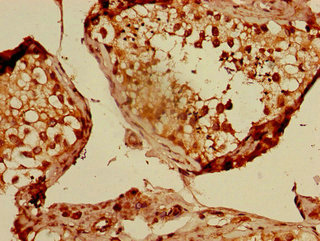
Core component of multiple cullin-RING-based ECS (ElonginB/C-CUL2/5-SOCS-box protein) E3 ubiquitin-protein ligase complexes, which mediate the ubiquitination of target proteins. CUL2 may serve as a rigid scaffold in the complex and may contribute to catalysis through positioning of the substrate and the ubiquitin-conjugating enzyme. The E3 ubiquitin-protein ligase activity of the complex is dependent on the neddylation of the cullin subunit and is inhibited by the association of the deneddylated cullin subunit with TIP120A/CAND1. The functional specificity of the ECS complex depends on the substrate recognition component. ECS(VHL) mediates the ubiquitination of hypoxia-inducible factor (HIF). A number of ECS complexes (containing either KLHDC2, KLHDC3, KLHDC10, APPBP2, FEM1A, FEM1B or FEM1C as substrate-recognition component) are part of the DesCEND (destruction via C-end degrons) pathway, which recognizes a C-degron located at the extreme C terminus of target proteins, leading to their ubiquitination and degradation. ECS complexes and ARIH1 collaborate in tandem to mediate ubiquitination of target proteins.