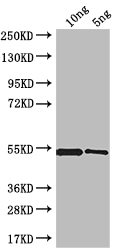
To generate polyclonal antibodies against MRC1, a rabbit is systematically immunized with recombinant Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) MRC1 protein (711-851aa). After reaching an optimal antibody titer, the rabbit is bled, and antibodies are extracted from the serum and then undergo affinity chromatography purification. The functionality of the resultant MRC1 antibody is then assessed in both ELISA and WB applications, demonstrating specificity towards Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) MRC1 protein.
The Saccharomyces cerevisiae MRC1 is involved in the regulation of DNA replication and the cellular response to DNA damage, thus maintaining chromosomal stability, preventing the accumulation of genetic mutations, and ensuring accurate transmission of genetic information during cell division in Saccharomyces cerevisiae. Essential for stabilizing and maintaining the integrity of the replication fork during DNA replication, MRC1 prevents replication fork collapse and facilitates smooth replication machinery progression. Additionally, MRC1 also participates in signaling pathways that mediate DNA repair and contribute to genomic integrity.