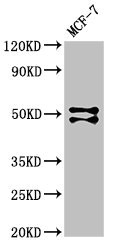
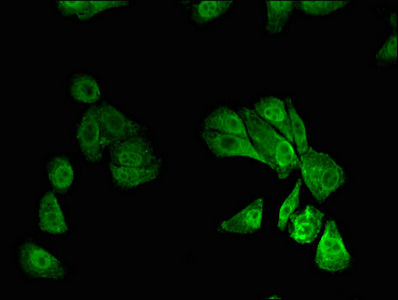
Forms the heterodimeric complex core-binding factor (CBF) with CBFB. RUNX members modulate the transcription of their target genes through recognizing the core consensus binding sequence 5'-TGTGGT-3', or very rarely, 5'-TGCGGT-3', within their regulatory regions via their runt domain, while CBFB is a non-DNA-binding regulatory subunit that allosterically enhances the sequence-specific DNA-binding capacity of RUNX. The heterodimers bind to the core site of a number of enhancers and promoters, including murine leukemia virus, polyomavirus enhancer, T-cell receptor enhancers, LCK, IL3 and GM-CSF promoters. Essential for the development of normal hematopoiesis. Acts synergistically with ELF4 to transactivate the IL-3 promoter and with ELF2 to transactivate the BLK promoter. Inhibits KAT6B-dependent transcriptional activation. Involved in lineage commitment of immature T cell precursors. CBF complexes repress ZBTB7B transcription factor during cytotoxic (CD8+) T cell development. They bind to RUNX-binding sequence within the ZBTB7B locus acting as transcriptional silencer and allowing for cytotoxic T cell differentiation. CBF complexes binding to the transcriptional silencer is essential for recruitment of nuclear protein complexes that catalyze epigenetic modifications to establish epigenetic ZBTB7B silencing. Controls the anergy and suppressive function of regulatory T-cells (Treg) by associating with FOXP3. Activates the expression of IL2 and IFNG and down-regulates the expression of TNFRSF18, IL2RA and CTLA4, in conventional T-cells. Positively regulates the expression of RORC in T-helper 17 cells.; Isoform AML-1G shows higher binding activities for target genes and binds TCR-beta-E2 and RAG-1 target site with threefold higher affinity than other isoforms. It is less effective in the context of neutrophil terminal differentiation.; Isoform AML-1L interferes with the transactivation activity of RUNX1.