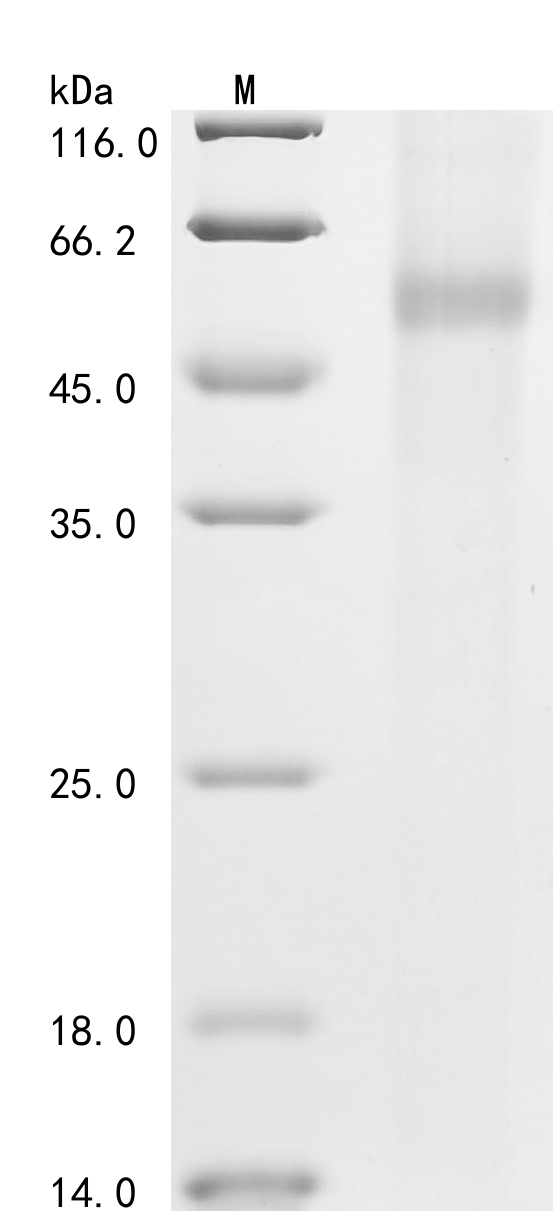
To produce recombinant human PARP1 in E. coli, the target gene is co-inserted into an expression plasmid with an N-terminal 6xHis-GST-tag gene and transformed into E. coli cells. The target gene codes for the 324-541aa of human PARP1. The cells are cultured to express the protein. The cells are lysed to release the recombinant PARP1 protein, which is purified using the affinity chromatography technique. Protein purity is evaluated by SDS-PAGE, reaching over 85%.
Human PARP1 is a DNA-dependent ADP-ribosylation transferase that has robust auto-PARylation activity, involving the addition of poly(ADP-ribose) chains to target proteins. It modulates protein ADP-ribosylation, affecting the functions of the modified proteins [1]. This enzyme is involved in genome stability, protein homeostasis, cell proliferation, differentiation, and apoptosis [2]. PARP1 regulates gene transcription and competes with histone deacetylases for the substrate NAD+ [3]. Activation of PARP1 is triggered by DNA damage, such as single-strand breaks, which can occur in response to inflammatory diseases, diabetes, reperfusion injury, or oxidative stress [4].
PARP1 is also involved in chromatin remodeling processes, contributing to local chromatin relaxation upon DNA damage [5]. Furthermore, PARP1 expression has been linked to various human cancers. Studies have shown that the upregulation of PARP1 in triple-negative breast cancer and endometrial cancer [6][7]. The research has found that PARP1 regulates the activity of influenza A virus polymerase, highlighting its role in viral replication processes [8].
References:
[1] H. Qi, Y. Lu, J. Lv, H. Wu, J. Lu, C. Zhanget al., The long noncoding rna lncparp1 contributes to progression of hepatocellular carcinoma through up-regulation of parp1, Bioscience Reports, vol. 38, no. 3, 2018. https://doi.org/10.1042/bsr20180703
[2] X. Zhang, A. Lam, Q. Cheng, V. Courouble, T. Strutzenberg, J. Liet al., Discovery of an nad+ analogue with enhanced specificity for parp1, Chemical Science, vol. 13, no. 7, p. 1982-1991, 2022. https://doi.org/10.1039/d1sc06256e
[3] A. Mangerich and A. Bürkle, Pleiotropic cellular functions of parp1 in longevity and aging: genome maintenance meets inflammation, Oxidative Medicine and Cellular Longevity, vol. 2012, p. 1-19, 2012. https://doi.org/10.1155/2012/321653
[4] S. Rajamohan, V. Pillai, M. Gupta, N. Sundaresan, K. Birukov, S. Samantet al., Sirt1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(adp-ribose) polymerase 1, Molecular and Cellular Biology, vol. 29, no. 15, p. 4116-4129, 2009. https://doi.org/10.1128/mcb.00121-09
[5] H. Sellou, T. Lebeaupin, C. Chapuis, R. Smith, A. Hegele, H. Singhet al., The poly(adp-ribose)-dependent chromatin remodeler alc1 induces local chromatin relaxation upon dna damage, Molecular Biology of the Cell, vol. 27, no. 24, p. 3791-3799, 2016. https://doi.org/10.1091/mbc.e16-05-0269
[6] V. Ossovskaya, I. Koo, E. Kaldjian, C. Alvares, & B. Sherman, Upregulation of poly (adp-ribose) polymerase-1 (parp1) in triple-negative breast cancer and other primary human tumor types, Genes & Cancer, vol. 1, no. 8, p. 812-821, 2010. https://doi.org/10.1177/1947601910383418
[7] F. Bi, D. Li, & Q. Yang, Hypomethylation of ets transcription factor binding sites and upregulation of parp1 expression in endometrial cancer, Biomed Research International, vol. 2013, p. 1-5, 2013. https://doi.org/10.1155/2013/946268
[8] L. Westera, A. Jennings, J. Maamary, M. Schwemmle, A. García‐Sastre, & E. Bortz, Poly-adp ribosyl polymerase 1 (parp1) regulates influenza a virus polymerase, Advances in Virology, vol. 2019, p. 1-11, 2019. https://doi.org/10.1155/2019/8512363