An organelle is a specialized subunit that performs specific functions within a cell. Organelles are embedded within the cytoplasm of eukaryotic and prokaryotic cells. In the more complex eukaryotic cells, organelles are often separately enclosed by their own lipid bilayers. The name organelle originates from the idea that these structures are parts of cells. Analogous to the body's internal organs, organelles are specialized and perform valuable functions necessary for normal cellular operation. Organelles have a wide range of responsibilities that include everything from generating energy for a cell to controlling the cell's growth and reproduction.
As shown in the Fig.1, these are several typical organelles in the animal cell. In this article, we review the function and special markers of these organelles respectively. Besides that, many organelles aren't present in the Fig.1, involving autophagosome, chromatin, cilia, endosome, exosome and melanosome.
Fig. 1. Components of a typical animal cell
Nucleolus
Both animal and plant cells contains a nucleolus, which is a small body in the nucleus of a cell that contains protein and RNA and is the site for the synthesis of ribosomal RNA and for the formation of ribosomal subunits. In cell biology, the nucleolus is defined as a sub-organelle of the nucleus of a cell (which itself is an organelle).
Structure
Nucleolus mainly consists of three major components - the dense fibrillar component (DFC), the fibrillar centers (FC) and granular components (GC)[1]. Nucleolus is a central nuclear location where all the ribosomal RNAs are processed, synthesized and collected with ribosomal proteins. Therefore, nucleolus is also known as the factory for ribosome production. Transcription of the rDNA occurs in the FC domain. The DFC contains the protein fibrillarin, which is important in rRNA processing. The GC contains the protein nucleophosmin, which is also involved in ribosome biogenesis[2].
Function and Markers
First, nucleolus, a non-membrane bound cellular structure, which is present within the nucleus of the cell and contains ribosomes having ribonucleic acid (RNA) in them, and plays a crucial role in regulating the transactions of proteins and also vitally modulating each and every cellular functions in the cell. Second, the main nucleolus function is to assemble and produce ribosome components within the cell. Besides that, nucleolus mainly acts as a suspension medium for cell-organelles in the nucleus of the cell.
Being one of the main sources of containing chromosomes with DNA present in it, nucleolus is highly rich in RNA and is used for maintaining a sound shape and structure of the nucleus. Other nucleolus functions like transportation of molecules, ions and vital substances for an effectual functioning of cell metabolism is included in the nucleolus functions as well.
In this part, we conclude several markers of nucleolus in the Table 1.
Table 1. The markers of nucleolus
|
organelle
|
Protein Name
|
Gene Name
|
Uniprot ID
|
|
nucleolus
|
DNA-directed RNA polymerase I subunit RPA34
|
CD3EAP
|
O15446
|
|
nucleolus
|
Eukaryotic translation initiation factor 6
|
EIF6
|
P56537
|
|
nucleolus
|
rRNA 2'-O-methyltransferase fibrillarin
|
FBL
|
P22087
|
|
nucleolus
|
Probable 28S rRNA (cytosine(4447)-C(5))-methyltransferase
|
NOP2
|
P46087
|
Nucleus
The cell nucleus is a membrane bound structure that contains the cell's hereditary information and controls the cell's growth and reproduction. It is the command center of a eukaryotic cell and is commonly the most prominent organelle in a cell.
Structure
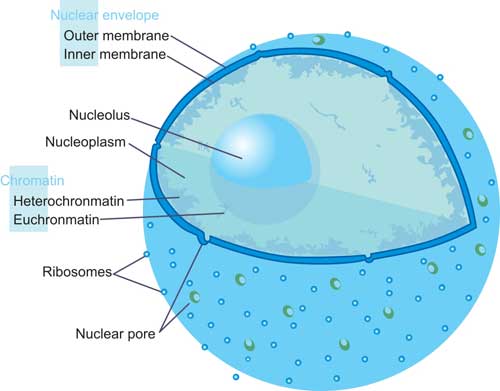
Fig.2. The Structure of Nucleus
Nucleus, the largest cellular organelle in animal cells, is bound by a double membrane called the nuclear envelope[3]. This membrane seems to be continuous with the endoplasmic reticulum (introduced in the next section in details) of the cell and has pores, which probably admit large molecules to enter. The average diameter of the nucleus in mammalian cells is approximately 6 micro-meters (µm), which occupies about 10% of the total cell volume. The viscous liquid within it is called nucleoplasm, and is similar in composition to the cytosol found outside the nucleus. It appears as a dense, roughly spherical or irregular organelle. The composition by dry weight of the nucleus is approximately: DNA 9%, RNA 1%, Histone Protein 11%, Residual Protein 14%, acidic Proteins 65%[4].
In some types of white blood cells specifically most granulocytes the nucleus is lobated and can be present as a bi-lobed, tri-lobed or multi-lobed organelle. As shown in the Fig.2.
Function and Markers
The nucleus provides a site for genetic transcription that is segregated from the location of translation in the cytoplasm, allowing levels of gene regulation that are not available to prokaryotes. The main function of the cell nucleus is to control gene expression and mediate the replication of DNA during the cell cycle.
In this section, we list several markers of nucleus in Table 2.
Table 2. The Markers of Nucleus
Ribosome
A tiny, somewhat mitten-shaped organelle occurring in great numbers in the cell cytoplasm either freely, in small clusters, or attached to the outer surfaces of endoplasmic reticula, and functioning as the site of protein manufacture.
Structure
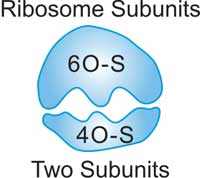
Fig.3. The Structure of Ribosome
Ribosomes are special because they are found in both prokaryotes and eukaryotes. There are two pieces or subunits to every ribosome. In eukaryotes, scientists have identified the 60-S (large) and 40-S (small) subunits. Even though ribosomes have slightly different structures in different species, their functional areas are all very similar. As shown in the Table 3, there are some difference of ribosomes between eukaryote and prokaryote.
Table 3. Compared the ribosome components of eukaryote with prokaryote
|
Ribosome
|
Subunit
|
rRNAs
|
r-proteins
|
|
eukaryote
|
80S
|
60S
|
28S (4718 nt)
|
49
|
|
5.8S (160 nt)
|
|
5S (120 nt)
|
|
40S
|
18S (1874 nt)
|
33
|
|
prokaryote
|
70S
|
50S
|
23S (2904 nt)
|
31
|
|
5S (120 nt)
|
|
30S
|
16S (1542 nt)
|
21
|
Function and Markers
Ribosomes are found in many places around a eukaryotic cell. You might find them floating in the cytosol. Those floating ribosomes make proteins that will be used inside of the cell. Other ribosomes are found on the endoplasmic reticulum. Endoplasmic reticulum with attached ribosomes is called rough ER. It looks bumpy under a microscope. The attached ribosomes make proteins that will be used inside the cell and proteins made for export out of the cell. There are also ribosomes attached to the nuclear envelope. Those ribosomes synthesize proteins that are released into the perinuclear space.
Ribosomes are a cell structure that makes protein. Protein is needed for many cell functions such as repairing damage or directing chemical processes. The location of the ribosomes in a cell determines what kind of protein it makes. If the ribosomes are floating freely throughout the cell, it will make proteins that will be utilized within the cell itself. When ribosomes are attached to endoplasmic reticulum, it is referred to as rough endoplasmic reticulum or rough ER. Proteins made on the rough ER are used for usage inside the cell or outside the cell.
In the following Table 4, we enumerate several markers in the research of ribosome.
Table 4. Markers of ribosome
Vesicle
A vesicle is a small membrane-bound sack that stores and transports substances throughout the cell. They can form naturally within the cell to aid in cellular secretion such as exocytosis, endocytosis or phagocytosis. Besides that, they also can be artificially prepared by chemists and biologists. In this article, we focus on two types of vesicle – Vacuole and lysosomes. The two types of vesicle will introduce in the later sections closely.
Endoplasmic reticulum
The endoplasmic reticulum (ER), an organelle, is an interconnected network of flattened sacs or tubes encased in membranes. These membranes are continuous, joining with the outer membrane of the nuclear membrane. All eukaryotic cells contain an ER, except red blood cells and sperm cells. Based on different physical and functional characteristics, the ER is divided into two types, known as rough ER and smooth ER.
Structure
The general structure of the endoplasmic reticulum is a network of membranes called cisternae. These sac-like structures are held together by the cytoskeleton. The phospholipid membrane encloses the cisternal space (or lumen), which is continuous with the perinuclear space but separate from the cytosol.
The functions of the endoplasmic reticulum can be summarized as the synthesis and export of proteins and membrane lipids, but varies between ER and cell type and cell function. The quantity of both rough and smooth endoplasmic reticulum in a cell can slowly interchange from one type to the other, depending on the changing metabolic activities of the cell. Transformation can include embedding of new proteins in membrane as well as structural changes. Changes in protein content may occur without noticeable structural changes.
Function and Markers
The rough endoplasmic reticulum manufactures membranes and secretory proteins. The ribosomes attached to the rough ER synthesize proteins by the process of translation. In certain leukocytes (white blood cells), the rough ER produces antibodies. In pancreatic cells, the rough ER produces insulin. The rough and smooth ER are usually interconnected and the proteins and membranes made by the rough ER move into the smooth ER to be transferred to other locations. Some proteins are sent to the Golgi apparatus by special transport vesicles. After the proteins have been modified in the Golgi, they are transported to their proper destinations within the cell or exported from the cell by exocytosis.
The smooth ER has a wide range of functions including carbohydrate and lipid synthesis. Lipids such as phospholipids and cholesterol are necessary for the construction of cell membranes. Smooth ER also serves as a transitional area for vesicles that transport ER products to various destinations. In liver cells the smooth ER produces enzymes that help to detoxify certain compounds. In muscles the smooth ER assists in the contraction of muscle cells, and in brain cells it synthesizes male and female hormones.
In the following Table 5, we enumerate several markers in the research of endoplasmic reticulum.
Table 5. Markers of Endoplasmic Reticulum
|
Organelle
|
Protein Name
|
Gene Name
|
Uniprot ID
|
|
Endoplasmic Reticulum
|
B-cell receptor-associated protein 31
|
BCAP31
|
P51572
|
|
Endoplasmic Reticulum
|
Calreticulin
|
CALR
|
P27797
|
|
Endoplasmic Reticulum
|
Calnexin
|
CANX
|
P27824
|
|
Endoplasmic Reticulum
|
Cytochrome P450 2E1
|
CYP2E1
|
P05181
|
|
Endoplasmic Reticulum
|
Heme oxygenase 1
|
HMOX1
|
P09601
|
|
Endoplasmic Reticulum
|
Heme oxygenase 2
|
HMOX2
|
P30519
|
|
Endoplasmic Reticulum
|
Endoplasmin
|
HSP90B1
|
P14625
|
|
Endoplasmic Reticulum
|
Endoplasmic reticulum chaperone BiP
|
HSPA5
|
P11021
|
|
Endoplasmic Reticulum
|
Protein disulfide-isomerase
|
P4HB
|
P07237
|
|
Endoplasmic Reticulum
|
Protein disulfide-isomerase A3
|
PDIA3
|
P30101
|
|
Endoplasmic Reticulum
|
Protein disulfide-isomerase A4
|
PDIA4
|
P13667
|
|
Endoplasmic Reticulum
|
Antigen peptide transporter 1
|
TAP1
|
Q03518
|
|
Endoplasmic Reticulum
|
Tapasin
|
TAPBP
|
O15533
|
|
Endoplasmic Reticulum
|
UDP-glucose:glycoprotein glucosyltransferase 1
|
UGGT1
|
Q9NYU2
|
|
Endoplasmic Reticulum
|
UDP-glucose:glycoprotein glucosyltransferase 2
|
UGGT2
|
Q9NYU1
|
Golgi apparatus
Golgi apparatus, also called Golgi complex or Golgi body, membrane-bound organelle of eukaryotic cells (cells with clearly defined nuclei) that is made up of a series of flattened, stacked pouches called cisternae.
Structure
The Golgi apparatus has a structure that is made up of cisternae, which are flattened stacks of membrane usually found in a series of five to eight, or until they fill up the cytoplasm. These cisternae help proteins and cytoplasmic components.
Function and Markers
The Golgi complex works closely with the rough ER. When a protein is made in the ER, something called a transition vesicle is made. This vesicle or sac floats through the cytoplasm to the Golgi apparatus and is absorbed. After the Golgi does its work on the molecules inside the sac, a secretory vesicle is created and released into the cytoplasm. From there, the vesicle moves to the cell membrane and the molecules are released out of the cell.
In the following Table 6, we enumerate several markers in the research of Golgi apparatus.
Table 6. Markers of Golgi Apparatus
|
Organelle
|
Protein Name
|
Gene Name
|
Uniprot ID
|
|
Golgi apparatus
|
Beta-1,4-galactosyltransferase 6
|
B4GALT6
|
Q9UBX8
|
|
Golgi apparatus
|
BET1 homolog
|
BET1
|
O15155
|
|
Golgi apparatus
|
Receptor-binding cancer antigen expressed on SiSo cells
|
EBAG9
|
O00559
|
|
Golgi apparatus
|
Formimidoyltransferase-cyclodeaminase
|
FTCD
|
O95954
|
|
Golgi apparatus
|
Golgin subfamily A member 1
|
GOLGA1
|
Q92805
|
|
Golgi apparatus
|
Golgin subfamily A member 2
|
GOLGA2
|
Q08379
|
|
Golgi apparatus
|
Golgin subfamily A member 5
|
GOLGA5
|
Q8TBA6
|
|
Golgi apparatus
|
Golgi reassembly-stacking protein 2
|
GORASP2
|
Q9H8Y8
|
|
Golgi apparatus
|
Golgi SNAP receptor complex member 1
|
GOSR1
|
O95249
|
|
Golgi apparatus
|
Golgi SNAP receptor complex member 2
|
GOSR2
|
O14653
|
|
Golgi apparatus
|
Alpha-mannosidase 2
|
MAN2A1
|
Q16706
|
|
Golgi apparatus
|
Lysosomal alpha-mannosidase
|
MAN2B1
|
O00754
|
|
Golgi apparatus
|
Beta-mannosidase
|
MANBA
|
O00462
|
|
Golgi apparatus
|
Syntaxin-6
|
STX6
|
O43752
|
|
Golgi apparatus
|
Trans-Golgi network integral membrane protein 2
|
TGOLN2
|
O43493
|
Cytoskeleton
A cytoskeleton is present in the cytoplasm of all cells, including bacteria and archaea. It is a complex, dynamic network of interlinking protein filaments that extends from the cell nucleus to the cell membrane. The cytoskeletal systems of different organisms are composed of similar proteins.
Structure
The cytoskeleton is composed of at least three different types of fibers: microtubules, microfilaments, and intermediate filaments. These fibers are distinguished by their size with microtubules being the thickest and microfilaments being the thinnest. In the Table 7 below, the difference among these three types of cytoskeleton are presented.
Table 7. Comparison of Three Types of Cytoskeleton
|
Cytoskeleton type
|
Diameter (nm)
|
Structure
|
Subunit examples
|
|
Microtubules
|
23
|
protofilaments, in turn consisting of tubulin subunits in complex with stathmin
|
α- and β-tubulin
|
|
Microfilaments
|
6
|
double helix
|
actin
|
|
Intermediate filaments
|
10
|
two anti-parallel helices/dimers, forming tetramers
|
vimentin (mesenchyme)
|
*the content of Table 7 is derived from Wikipedia.
Function and Markers
The cytoskeleton extends throughout the cell's cytoplasm and directs a number of important functions, involving the maintenance of cell shape, held variety of cellular organelles in their place, and assistance of vacuoles formation. Note that, the cytoskeleton is not a static structure, but is able to disassemble and reassemble its parts in order to enable internal and overall cell mobility.
In the following Table 8, we enumerate several markers in the research of cytoskeleton.
Table 8. Markers of Cytoskeleton
|
Organelle
|
Protein Name
|
Gene Name
|
Uniprot ID
|
|
cytoskeleton
|
Actin, cytoplasmic 1
|
ACTB
|
P60709
|
|
cytoskeleton
|
Desmin
|
DES
|
P17661
|
|
cytoskeleton
|
Keratin, type I cytoskeletal 17
|
KRT17
|
Q04695
|
|
cytoskeleton
|
Keratin, type I cytoskeletal 20
|
KRT20
|
P35900
|
|
cytoskeleton
|
Keratin, type II cytoskeletal 7
|
KRT7
|
P08729
|
|
cytoskeleton
|
Neuroendocrine convertase 1
|
PCSK1
|
P29120
|
|
cytoskeleton
|
Tubulin alpha-1A chain
|
TUBA1A
|
Q71U36
|
|
cytoskeleton
|
Tubulin beta chain
|
TUBB
|
P07437
|
|
cytoskeleton
|
vimentin
|
VIM
|
P08670
|
Mitochondrion
Mitochondrion, membrane-bound organelle found in the cytoplasm of almost all eukaryotic cells (cells with clearly defined nuclei), the primary function of which is to generate large quantities of energy in the form of adenosine triphosphate (ATP).
Structure
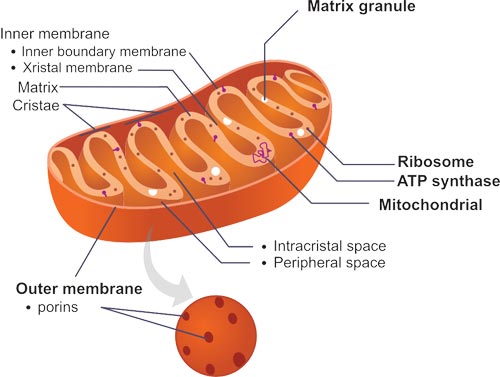
Fig.4. The Structure of Mitochondria
Mitochondria are typically round to oval in shape and range in size from 0.5 to 10 μm, and contains outer and inner membranes composed of phospholipid bilayers and proteins. Due to the different properties of outer and inner membranes, there are five distinct sections to a mitochondrion- the outer mitochondrial membrane, the inter-membrane space (the space between the outer and inner membranes), the inner mitochondrial membrane, the cristae space (formed by infoldings of the inner membrane), and the matrix (space within the inner membrane).
Function and Markers
The primary function of which is to generate large quantities of energy in the form of adenosine triphosphate (ATP), which is a series of reactions (also known as the citric acid cycle, or the Krebs cycle). In addition to producing energy, mitochondria store calcium for cell signaling activities, generate heat, and mediate cell growth and death. The number of mitochondria per cell varies widely; for example, in humans, erythrocytes (red blood cells) do not contain any mitochondria, whereas liver cells and muscle cells may contain hundreds or even thousands. The only eukaryotic organism known to lack mitochondria is the oxymonad Monocercomonoides species.
In the following Table 9, we enumerate several markers in the research of mitochondrion.
Table 9. Markers of Mitochondrion
|
Organellef
|
Protein Name
|
Gene Name
|
Uniprot ID
|
|
Mitochondria
|
Apoptosis-inducing factor 1, mitochondrial
|
AIFM1
|
O95831
|
|
Mitochondria
|
Cytochrome c oxidase subunit 4 isoform 1, mitochondrial
|
COX4I1
|
P13073
|
|
Mitochondria
|
Carbamoyl-phosphate synthase [ammonia], mitochondrial
|
CPS1
|
P31327
|
|
Mitochondria
|
FAST kinase domain-containing protein 2, mitochondrial
|
FASTKD2
|
Q9NYY8
|
|
Mitochondria
|
Hexokinase-1
|
HK1
|
P19367
|
|
Mitochondria
|
Heat shock 70 kDa protein 1A
|
HSPA1A
|
P0DMV8
|
|
Mitochondria
|
60 kDa heat shock protein, mitochondrial
|
HSPD1
|
P10809
|
|
Mitochondria
|
Mitochondrial dynamics protein MID51
|
MIEF1
|
Q9NQG6
|
|
Mitochondria
|
Cytochrome c oxidase subunit 1
|
MT-CO1
|
P00395
|
|
Mitochondria
|
NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial
|
NDUFS7
|
O75251
|
|
Mitochondria
|
Mitochondrial import inner membrane translocase subunit TIM16
|
PAM16
|
Q9Y3D7
|
|
Mitochondria
|
Prohibitin
|
PHB
|
P35232
|
|
Mitochondria
|
Superoxide dismutase [Cu-Zn]
|
SOD1
|
P00441
|
|
Mitochondria
|
Mitochondrial import receptor subunit TOM22 homolog
|
TOMM22
|
Q9NS69
|
|
Mitochondria
|
Elongation factor Tu, mitochondrial
|
TUFM
|
P49411
|
|
Mitochondria
|
Mitochondrial brown fat uncoupling protein 1
|
UCP1
|
P25874
|
|
Mitochondria
|
Mitochondrial uncoupling protein 2
|
UCP2
|
P55851
|
|
Mitochondria
|
Mitochondrial uncoupling protein 3
|
UCP3
|
P55916
|
|
Mitochondria
|
Voltage-dependent anion-selective channel protein 1
|
VDAC1
|
P21796
|
Vacuole
A vacuole is a cell organelle found in a number of different cell types. Vacuoles are fluid-filled, enclosed structures that are separated from the cytoplasm by a single membrane. They are found mostly in plant cells and fungi. However, some protists, animal cells, and bacteria also contain vacuoles. Vacuoles are responsible for a wide variety of important functions in a cell including nutrient storage, detoxification, and waste exportation.
Cytosol
Cytosol is the intra-cellular fluid that is present inside living cells. More specifically, cytosol mainly consists of water, which occupies 70% of the total volume of a cell. The cytosol has no single function and is instead the site of multiple cell processes. Examples of these processes include signal transduction from the cell membrane to sites within the cell, such as the cell nucleus, or organelles. Sometimes, cytosol is often confused with cytoplasm. But, in fact, they are entirely different entities within a cell. Cytosol comprises of water and anything soluble that is dissolved in it, such as soluble proteins and ions. However, the cytoplasm consists of cytosol and certain insoluble suspended particles, such as ribosomes.
Lysosome
Lysosome, subcellular organelle that is found in nearly all types of eukaryotic cells (cells with a clearly defined nucleus) and that is responsible for the digestion of macromolecules, old cell parts, and microorganisms. Lysosomes were discovered by the Belgian cytologist Christian Rene de Duve in the 1950s.
Structure and function
Each lysosome is surrounded by a membrane that maintains an acidic environment within the interior via a proton pump. Lysosomes contain a wide variety of hydrolytic enzymes (acid hydrolases) that break down various macromolecules it engulfs, invovling nucleic acids, proteins, and polysaccharides. These enzymes are active only in the lysosome's acidic interior; their acid-dependent activity protects the cell from self-degradation in case of lysosomal leakage or rupture, since the pH of the cell is neutral to slightly alkaline. In addition to being able to break down polymers, lysosomes are capable of fusing with other organelles & digesting large structures or cellular debris; through cooperation with phagosomes, they are able to conduct autophagy, clearing out damaged structures. Similarly, they are able to break-down virus particles or bacteria in phagocytosis of macrophages.
Markers
In the following Table 10, we enumerate several markers in the research of lysosome. We hope that will give some help for you.
Table 10. Markers of Lysosome
|
Organelle
|
Protein Name
|
Gene Name
|
Uniprot ID
|
|
Lysosome
|
Ubiquitin-like protein ATG12
|
ATG12
|
O94817
|
|
Lysosome
|
Autophagy protein 5
|
ATG5
|
Q9H1Y0
|
|
Lysosome
|
Beta-galactosidase
|
GLB1
|
P16278
|
|
Lysosome
|
Integral membrane protein
GPR137B
|
GPR137B
|
O60478
|
|
Lysosome
|
Lysosome-associated membrane glycoprotein 1
|
LAMP1
|
P11279
|
|
Lysosome
|
Lysosome-associated membrane glycoprotein 2
|
LAMP2
|
P13473
|
|
Lysosome
|
Microtubule-associated proteins 1A/1B light chain 3A
|
MAP1LC3A
|
Q9H492
|
|
Lysosome
|
Rap1 GTPase-activating protein 1
|
RAP1GAP
|
P47736
|
Centrosome
Centrosome is an organelle near the nucleus in the cytoplasm, which contains the centrioles (in animal cells) and from which the spindle fibers develop in cell division. During mitosis, centrosome divides into two parts and migrates to opposite poles of the cell, and is involved in the formation of mitotic spindle, assembly of microtubules, and regulation of cell cycle progression.
Structure
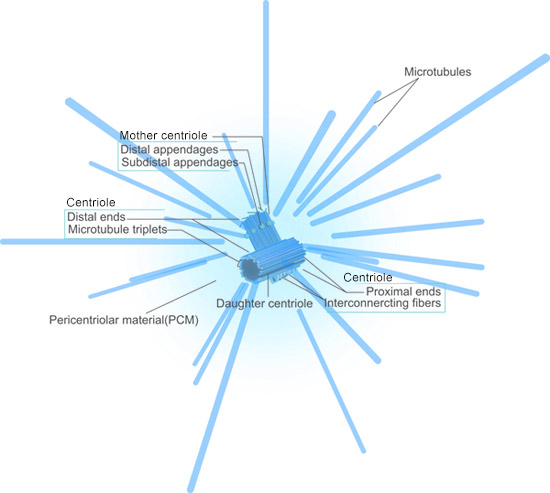
Fig.4. The Structure of Centrosome
The animal centrosome consists of a pair of centrioles linked together with their proximal regions. The centrioles contain cylindrical arrays of triplet MTs organized with nine-fold radial symmetry, and the proximal region is structurally similar to the basal bodies of cilia and flagella[5][6][7].
In post-mitotic cells, the centrosome contains a mature centriole called the mother centriole and an immature centriole assembled in the previous cell cycle, the daughter centriole, which is about 80% the length of the mother centriole[8]. Mother centrioles are distinguished by two sets of nine appendages at their distal ends[9].
Function and Markers
The function of centrosome, during cell division, is to maintain the chromosome number. Replication of DNA is accompanied by the splitting of the centrosome into two, each having one centriole during cell division. The two centrosomes attach themselves to the opposite poles of the nucleus. Sometimes, you may confuse with the concepts of centrosome and chromosome. Actually, as the nouns mentioned, centrosome is an organelle, near the nucleus in the cytoplasm of most organisms that controls the organization of its microtubules. While chromosome is a structure in the cell nucleus that contains DNA, histone protein, and other structural proteins.
In the following Table 11, we list several markers in the research of centrosome. We hope that will give some help for you.
Table 11. The Markers of Centrosome
Cell membrane
The cell membrane, also known as the plasma membrane or cytoplasmic membrane, a thin semi-permeable membrane that surrounds the cytoplasm of a cell and separates the interior of all cells from the outside environment (the extracellular space).
Structure and function
The cell membrane is primarily composed of a mix of proteins and lipids. Depending on the membrane's location and role in the body, lipids can make up anywhere from 20 to 80 percent of the membrane with the remainder being proteins. Besides that, lipids make membranes flexible, proteins monitor and maintain the cell's chemical climate and assist in the transfer of molecules across the membrane.
The main function of cell membrane is to protect the integrity of the interior of the cell via allowing certain substances into the cell while keeping other substances out. It also serves as a base of attachment for the cytoskeleton in some organisms and the cell wall in others. Thus the cell membrane also serves to help support the cell and help maintain its shape.
Another function of the membrane is to regulate cell growth via the balance of endocytosis and exocytosis. In endocytosis, vesicles containing lipids and proteins fuse with the cell membrane increasing cell size as internalized substances. In exocytosis, vesicles containing lipids and proteins lipids and proteins are removed from the cell membrane. Animal cells, plant cells, prokaryotic cells, and fungal cells have plasma membranes. Internal organelles are also encased by membranes.
Markers
In the following Table 12, we list a number of markers of cell membrane, we hope that will help your research.
Table 12. The Markers of Cell Membrane
|
Organelle
|
Protein Name
|
Gene Name
|
Uniprot ID
|
|
Cell membrane
|
CD27 antigen
|
CD27
|
P26842
|
|
Cell membrane
|
CD40 ligand
|
CD40LG
|
P29965
|
|
Cell membrane
|
Cadherin EGF LAG seven-pass G-type receptor 1
|
CELSR1
|
Q9NYQ6
|
|
Cell membrane
|
Death domain-containing protein CRADD
|
CRADD
|
P78560
|
|
Cell membrane
|
Ezrin
|
EZR
|
P15311
|
|
Cell membrane
|
FAS-associated death domain protein
|
FADD
|
Q13158
|
|
Cell membrane
|
Tumor necrosis factor ligand superfamily member 6
|
FASLG
|
P48023
|
|
Cell membrane
|
Growth factor receptor-bound protein 2
|
GRB2
|
P62993
|
|
Cell membrane
|
Merlin
|
NF2
|
P35240
|
|
Cell membrane
|
Tumor necrosis factor receptor superfamily member 1A
|
TNFRSF1A
|
P19438
|
Chromatin
Chromatin is a mass of genetic material composed of DNA, proteins and RNA that condense to form chromosomes during eukaryotic cell division. Chromatin is located in the nucleus of our cells.
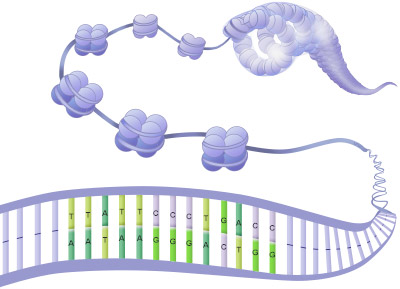
Fig.5. The Structure of Chromatin
Comparison
Concerning the study of chromatin, we always mix the concepts of chromatin and chromosome. In this section, we compare the two things from below aspects (as shown in the Table 13).
Table 13. The Comparison between Chromatin and Chromosome
|
Chromatin
|
Chromosome
|
|
Definition
|
In the nucleus, the DNA double helix is packaged by special proteins (histones) to form a complex called chromatin. The chromatin undergoes further condensation to form the chromosome.
|
A compact structure of nucleic acids and protein found in the nucleus of most living cells, carrying genetic information in the form of genes.
|
|
Structure
|
Composed of nucleosomes-a complex of DNA and proteins (called histones). Represent DNA folded on nucleoproteins by a magnitude of 50. The chromatin fiber is app. 10 nm in diameter.
|
Chromosomes are condensed Chromatin Fibers. They are a higher order of DNA organization, where DNA is condensed at least by 10,000 times onto itself.
|
|
Appearance
|
Chromatin Fibers are Long and thin. They are uncoiled structures found inside the nucleus.
|
Chromosomes are compact, thick and ribbon-like. These are coiled structures seen prominently during cell division.
|
|
Pairs
|
Chromatin is unpaired.
|
Chromosome is paired.
|
|
Metabolic activity
|
Permissive to DNA replication, RNA synthesis (transcription) and recombination events.
|
Refractory to these processes.
|
|
Presence
|
Found throughout the cell cycle.
|
Distinctly visible during cell division (metaphase, anaphase) as highly condensed structures upto several thousand nm.
|
|
Conformation
|
May have open (euchromatin) or compact (heterochromatin) conformations, which is dynamically regulated during the cell-cycle stages.
|
Predominantly heterochromatic state with a predetermined position in the nucleus and a specific shape such as metacentric, submetacentric, acrocentric, telocentric.
|
|
Visualization
|
Electron microscope (beads on string appearance)
|
Light microscope (classic four-arm structure when duplicated)
|
Function and Markers
The main function of chromatin is to condense the DNA into a compact unit that will be less voluminous and can fit within the nucleus. Chromatin consists of complexes of histones and DNA. Histones help to organize DNA into structures called nucleosomes by providing a base on which the DNA can be wrapped around. A nucleosome consists of a DNA sequence of about 150 base pairs that is wrapped around a set of eight histones called an octamer. The nucleosome is further folded to produce a chromatin fiber. Chromatin fibers are coiled and condensed to form chromosomes. Chromatin makes it possible for a number of cell processes to occur including DNA replication, transcription, DNA repair, genetic recombination, and cell division.
In the following Table 13, we list a number of markers of chromatin, we hope that will help your research.
Table 13. The Markers of Chromatin
Autophagosome
An autophagosome is a spherical structure with double layer membranes. It is a central structure in macroautophagy, the system of intracellular degradation for cytoplasmic contents and also for invading microorganisms. After formation, autophagosomes deliver cytoplasmic components to the lysosomes. If you want to read more information about autophagy, please click the below link: https://www.cusabio.com/c-20666.html
In the following Table 14, we list several autophagosome markers, which can aid in the study of the structure, function, and formation of the autophagosome.
Table 14. The Markers of Autophagosome
|
Organelle
|
Protein Name
|
Gene Name
|
Uniprot ID
|
|
Autophagosome
|
Microtubule-associated proteins 1A/1B light chain 3B
|
MAP1LC3B
|
Q9GZQ8
|
|
Autophagosome
|
Sequestosome-1
|
SQSTM1
|
Q13501
|
Cilium
Cilium (the plural is cilia) is microscopic, hair-like structure that extend outward from the surface of many animal cells.
Structure and function
The length of a typical cilium is 1~10μm, and the width of that is usually less than 1μm. They are often divided into two different types – motile and non-motile, and these types can work together or separately.
Motile cilia, as named moving, can be found in the respiratory tract, middle ear, and other body systems. Multiple cilia will wave in a rhythmic or pulsating motion, and use that motion to keep sensitive internal passage ways free of mucus or foreign particles, for example. Body cells that have a single moving cilium are sperm cells, which use that cilium to propel the cell.
The non-motile cilia play a key role in many different organs. Some almost act as antenna that receive sensory information for the cell, processing signals from the other cells or the fluids surrounding it. For example, cilia in the kidney are forced to bend as urine flows past, which sends signals to the cells that it is flowing. Non-motile cilia inside the eye are housed in the retina's photoreceptors, allowing important molecules to be transported from one end to the other.
Markers
In the following Table 14, we list a number of markers of cilium, we hope that will help your research.
Table 14. The Markers of Cilium
|
Organelle
|
Protein Name
|
Gene Name
|
Uniprot ID
|
|
Cilium
|
ADP-ribosylation factor-like protein 13B
|
ARL13B
|
Q3SXY8
|
|
Cilium
|
Intraflagellar transport protein 88 homolog
|
IFT88
|
Q13099
|
|
Cilium
|
Ras-related protein Rab-23
|
RAB23
|
Q9ULC3
|
Endosome
Endosome, formed by a complex family of processes collectively known as endocytosis, is a membrane-bound vesicle. It can be found in the cytoplasm of virtually every animal cell. The basic mechanism of endocytosis is the reverse of what occurs during exocytosis or cellular secretion. Endosomes comprise three different compartments, including early endosomes, late endosomes, and recycling endosomes[10]. They are distinguished through the time it takes for endocytosed material to reach them. They also have different morphology. Once endocytic vesicles have uncoated, they fuse with early endosomes. Early endosomes then mature into late endosomes before fusing with lysosomes[11][12][13].
Here, we numerate several markers of endosome, hope that will help your research.
Table 15. The Markers of Endosome
Exosome
Exosome, described as such 30 years ago firstly, is released from cells upon fusion of an intermediate endocytic compartment, the multivesicular body (MVB), with the plasma membrane[14]. If you want to know the exosome better, please click here.
In the following Table 16, there are many markers of exosome for you.
Table 16. The Markers of Exosome
Peroxisome
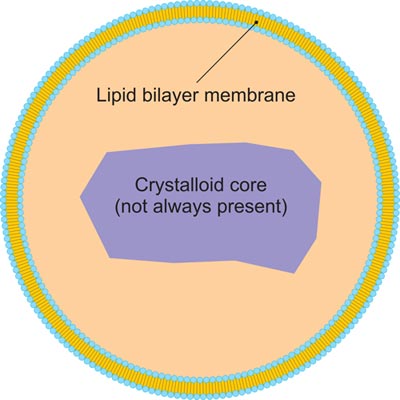
Fig.6. The structure of peroxisome
Peroxisome, a membrane-bound organelle occurring in the cytoplasm of eukaryotic cells, contains enzymes that oxidize certain molecules normally found in the cell, notably fatty acids and amino acids. Those oxidation reactions produce hydrogen peroxide, which is the basis of the name peroxisome. However, hydrogen peroxide is potentially toxic to the cell, because it has the ability to react with many other molecules. Therefore, peroxisomes also contain enzymes such as catalase that convert hydrogen peroxide to water and oxygen, thereby neutralizing the toxicity. In that way peroxisomes provide a safe location for the oxidative metabolism of certain molecules. Peroxisomes play a key role in the oxidation of specific biomolecules. They also contribute to the biosynthesis of membrane lipids known as plasmalogens. In plant cells, peroxisomes carry out additional functions, including the recycling of carbon from phosphoglycolate during photorespiration. Specialized types of peroxisomes have been identified in plants, among them the glyoxysome, which functions in the conversion of fatty acids to carbohydrates.
Here, we numerate several markers of peroxisome in the Table 17, hope that will help your research.
Table 17. The Markers of Peroxisome
|
Organelle
|
Protein Name
|
Gene Name
|
Uniprot ID
|
|
Peroxisome
|
ATP-binding cassette sub-family D member 4
|
ABCD4
|
O14678
|
|
Peroxisome
|
Catalase
|
CAT
|
P04040
|
|
Peroxisome
|
Matrix metalloproteinase-9
|
MMP9
|
P14780
|
|
Peroxisome
|
ATP-binding cassette sub-family D member 3
|
ABCD3
|
P28288
|
|
Peroxisome
|
Acyl-coenzyme A thioesterase 8
|
ACOT8
|
O14734
|
|
Peroxisome
|
Peroxisome biogenesis factor 1
|
PEX1
|
O43933
|
|
Peroxisome
|
Peroxisomal biogenesis factor 3
|
PEX3
|
P56589
|
|
Peroxisome
|
Peroxisomal targeting signal 1 receptor
|
PEX5
|
P50542
|
References
[1] O'Sullivan JM, Pai DA, et al. The nucleolus: a raft adrift in the nuclear sea or the keystone in nuclear structure? [J]. Biomolecular Concepts. 2013, 4 (3): 277–86.
[2] Sirri V, Urcuqui-Inchima S, et al. Nucleolus: the fascinating nuclear body [J]. Histochemistry and Cell Biology. 2008, 129 (1): 13–31.
[3] Lodish, H; Berk A; et al. Molecular Cell Biology (5th ed.). New York: WH Freeman. 2004, ISBN 0-7167-2672-6.
[4] Clegg JS. Properties and metabolism of the aqueous cytoplasm and its boundaries [J]. Am. J. Physiol. 1984, 246 (2 Pt 2): R133–51.
[5] Juliette Azimzadeh, Michel Bornens. Structure and duplication of the centrosome [J]. Journal of Cell Science 2007, 120: 2139-2142.
[6] Dawe, H. R., Farr, H. et al. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells [J]. 2007, J. Cell Sci. 120, 7-15.
[7] Davis, E. E., Brueckner, M. et al. The emerging complexity of the vertebrate cilium: new functional roles for an ancient organelle [J]. 2006, Dev. Cell 11, 9-19.
[8] Chretien, D., Buendia, B., et al. Reconstruction of the centrosome cycle from cryoelectron micrographs [J]. J. Struct. Biol. 1997, 120, 117-133.
[9] Paintrand, M., Moudjou, M., et al. Centrosome organization and centriole architecture: their sensitivity to divalent cations [J]. J. Struct. Biol. 1992, 108, 107-128.
[10] Mellman I. Endocytosis and molecular sorting [J]. Annual Review of Cell and Developmental Biology. 1996, 12: 575–625.
[11] Stenmark, H. Rab GTPases as coordinators of vesicle traffic [J]. Nat Rev Mol Cell Biol. 2009, 10 (8): 513–25.
[12] Futter, CE, Pearse, A.;et al. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes [J]. J Cell Biol. 1996, 132 (6): 1011–23.
[13] Luzio JP; Rous BA; et al. Lysosome-endosome fusion and lysosome biogenesis [J]. Journal of Cell Science. 2000, 113: 1515–1524.
[14] James R. Edgar. Q&A: What are exosomes, exactly [J]? BMC Biol. 2016, 14 (46): 2-7.
CUSABIO team. The Overview of Organelles. https://www.cusabio.com/c-20731.html










Comments
Leave a Comment