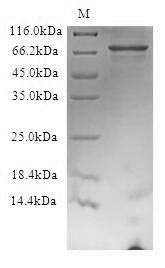
The production of this Recombinant Human FBXW7 protein required the insertion of a DNA fragment (FBXW7, 1-707aa) into an expression vector (a plasmid vector) and the transferral of this vector into in vitro E.coli cells (the step of transformation). The cells were then cultured and induced to express the FBXW7 protein. This recombinant protein was fused with N-terminal 6xHis tag. Its purity is 90%+ determined by SDS-PAGE.
FBXW7 functions as the substrate recognition component of the SCF E3 ubiquitin ligase. It is a key tumor suppressor and one of the most frequently deregulated ubiquitin-proteasome system proteins in human cancer. It specifically controls proteasomal degradation of oncoproteins such as cyclin E, c-Myc, Mcl-1, mTOR, Jun, Notch, and AURKA. FBXW7 gene mutations or decreased expression have a crucial biological function in tumor development. FBXW7 is also involved in embryonic development, as well as the proliferation and maturation of stem cell populations.